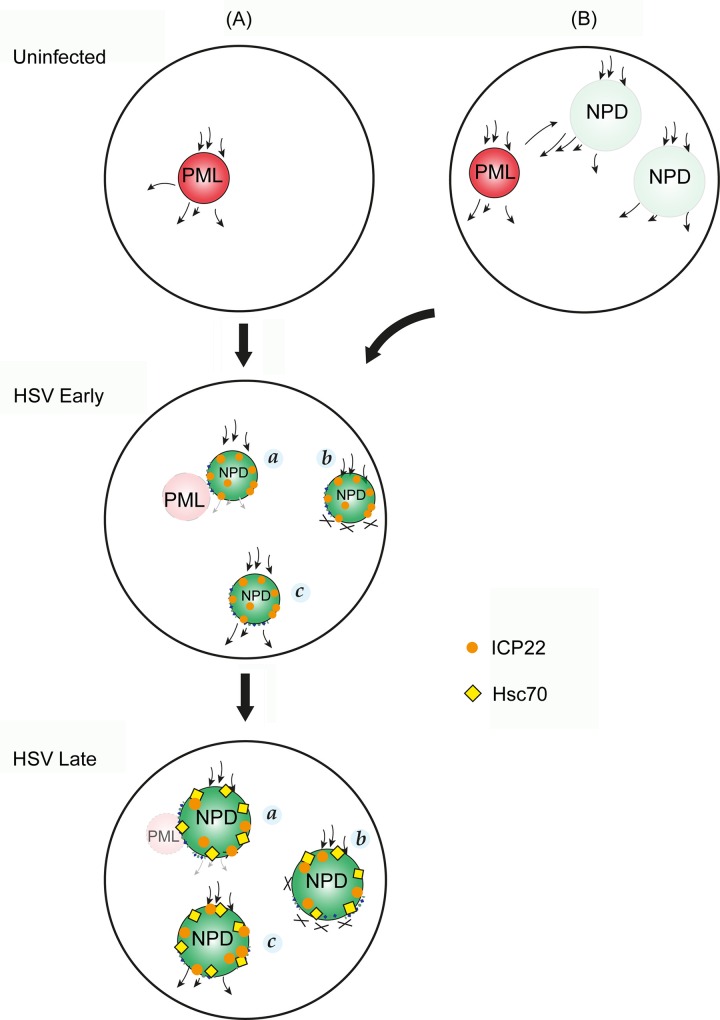
Fig 15. Schematic model of the spatial relationship between NPDs, PML domains and VICE domains and potential processing pathways of newly synthesised proteins.
The dark red shading indicates PML domains in uninfected cells and their functioning with dynamic recruitment and dissociation of target proteins (arrows in and out). The transparent lighter red shading in HSV infected cells indicates their progressive structural and functional disruption. NPDs are formed de novo only after infection (A) or in model (B), the possibility of pre-existing functional NPDs is indicated. Like PML domains, these could represent sites of dynamic protein recruitment and onward transport. As such they would not accumulate bulk newly synthesised proteins, and are thus indicated with transparent, light green shading. Their functional disruption, or overload, in infected cells is indicated by the dark green shading, firstly as smaller domains recruiting selectively ICP22, and only later in infection recruiting additional proteins including components of the host cell protein quality control machinery and in particular Hsc70, the diagnostic marker of VICE domain formation. As discussed in the text, small arrows (NPD,a) indicate impaired forward transport from NPDs and crosses (NPD,b) indicating a more complete block of transport of client proteins after continual recruitment. NPD,c, indicates a more pronounced forward transport after recruitment of certain classes of client proteins. ICP22 (magenta sphere), but not Hsc70, are recruited to NPDs during early stage of infection. As infection progresses, bulk Hsc70 (yellow diamond), and polyubiquitinated species together with ICP22, accumulates at NPDs of larger size.