Figure 1.
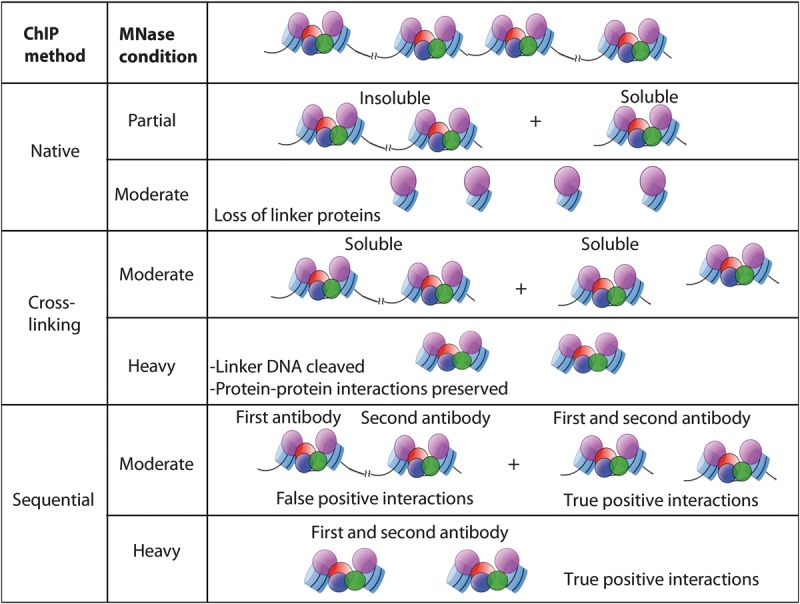
Schematic representation of comparative ChIP methods. Under native conditions, partial MNase digestion produces insoluble chromatin arrays as the major population. Upon moderate MNase digestion under similar conditions, only nucleosomes or similar structures that wrap DNA are protected; and therefore, proteins associated with the linker DNA are lost from the chromatin. In X-ChIP and sequential ChIP, however, protein–protein interactions are stabilized by cross-linking, and the solubility of chromatin is enhanced by detergents, resulting in ChIP signals after both moderate and heavy MNase digestion. In sequential ChIP, false positives can arise from there being multiple complexes pulled down from longer arrays, in which the first antibody pulls down one complex and the second antibody pulls down a different complex: (DNA) black line; (nucleosomes) blue barrels; (DNA-binding proteins) colored balls.
