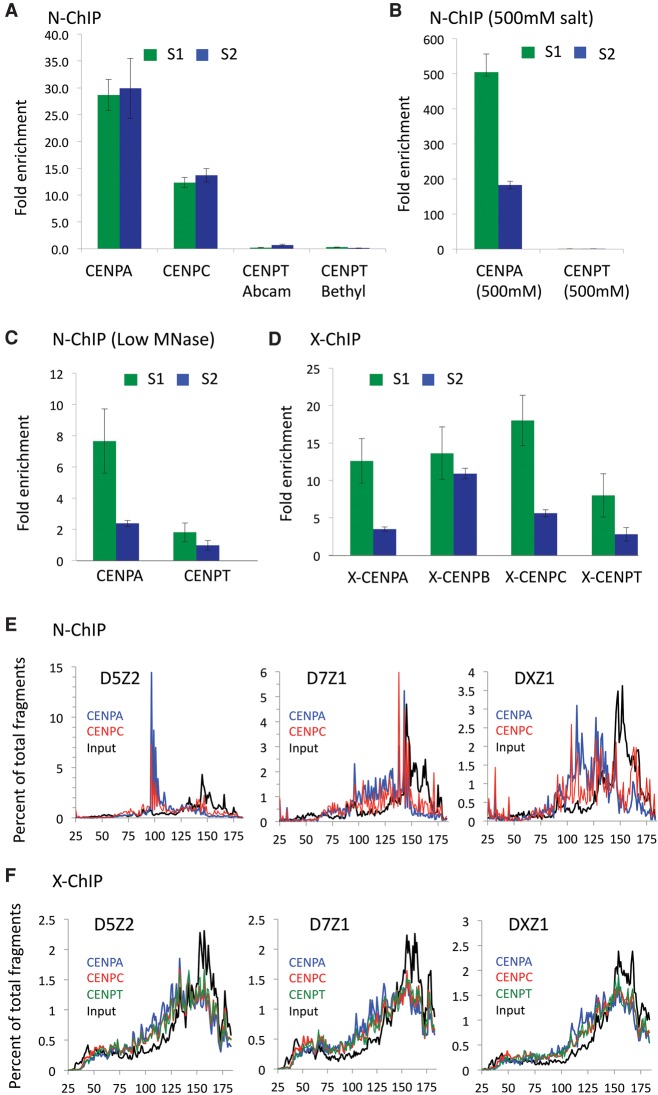
Figure 2.
CENPT is not retained at α-satellites by N-ChIP but is retained by X-ChIP. (A) Real-time PCR analysis of DNA obtained by native ChIP of CENPA, CENPC, and CENPT (using two different antibodies). The S1 primer pair was designed from Chromosome 21 alphoid arrays but is also present on Chromosome 13. Primer pair S2 was designed from the Cen1-like (SF1 family) sequences and is also present on Chromosomes 5 and 19 (Supplemental Table 1). Fold-enrichment over input was calculated relative to a control noncentromeric sequence from the 5S rDNA locus: Centromere (ChIP/input) ÷ 5S rDNA (ChIP/input). (B) CENPA and CENPT enrichment from native ChIP assays performed under 500 mM salt conditions. (C) CENPA and CENPT enrichment on DNA obtained from native ChIP assays performed on partially digested chromatin. (D) Enrichment of centromeric sequences in CENPA, CENPC, and CENPT cross-linking ChIP (X-ChIP). (E,F) Fragment length analysis of merged pairs obtained from (E) native or (F) X-ChIP data sets on D5Z2 (left), D7Z1 (middle), and DXZ1 (right) sequences. The sharp reduction in the size distribution above ∼160 bp and truncation at 185 bp is attributable to mapping of only paired-end reads in which the two 100-bp reads in a pair overlapped by 15 bp.