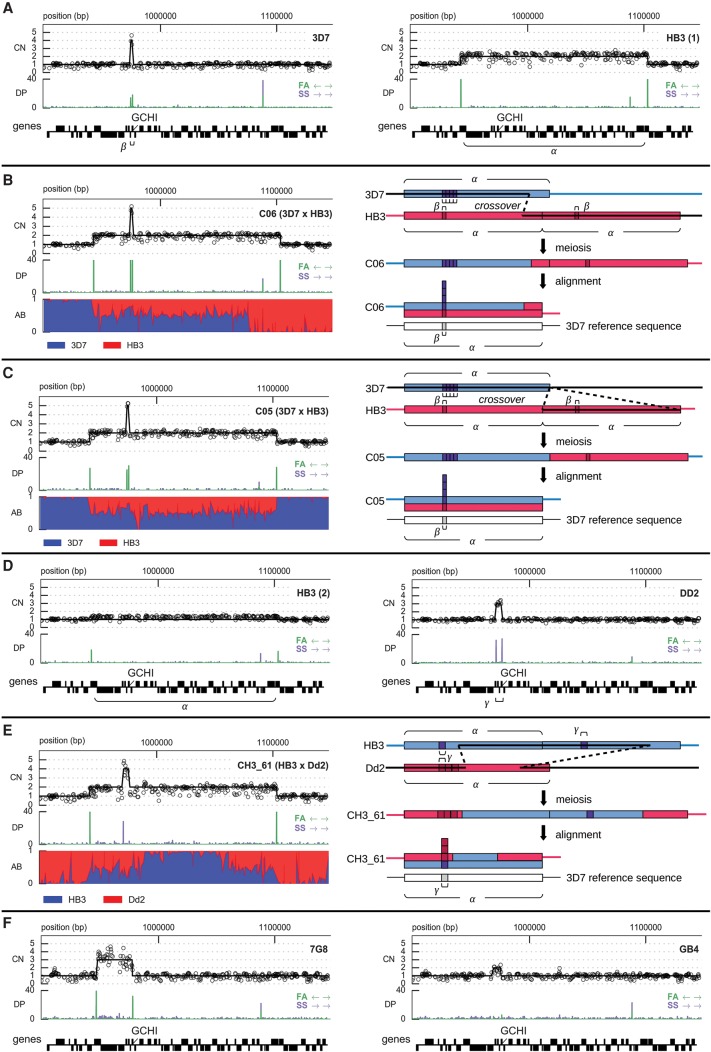
Figure 4.
Copy number variation and recombination spanning the anti-folate resistance gene gch1 on Chromosome 12. (A) CNVs in the 3D7 and HB3(1) parental clones; α labels the segment amplified in HB3, β labels the segment amplified in 3D7. (B) CNV and recombination in clone C06, progeny of 3D7 × HB3. AB = fraction of aligned reads containing the first parent's allele. (C) CNV and recombination in clone C05, progeny of 3D7 × HB3. AB = fraction of aligned reads containing the first parent's allele. (D) CNVs in the HB3(2) and Dd2 parental clones; γ labels the segment amplified in Dd2. Note that the HB3(2) clone sequenced here appears to be a mixture, with a minor proportion of parasites carrying the amplification visible in HB3(1). (E) CNV and recombination in clone CH3_61, progeny of HB3 × Dd2. AB = fraction of aligned reads containing the first parent's allele. (F) CNVs in the 7G8 and GB4 parental clones. CN = copy number; markers show normalized read counts within 300-bp nonoverlapping windows, excluding windows where GC content was below 20%; solid black line is the copy number predicted by fitting a Gaussian hidden Markov model to the coverage data (Supplemental Information). DP = depth of coverage (number of aligned reads), FA = reads aligned facing away from each other (expected at boundaries of a tandem array), SS = reads aligned in the same orientation (expected at boundaries of a tandem inversion).