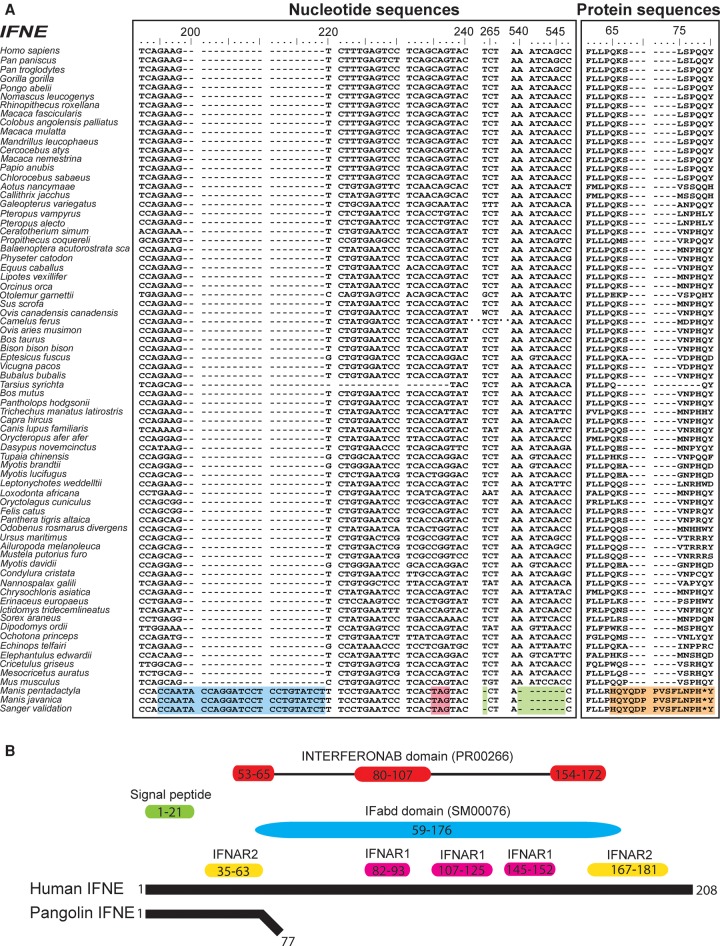
Figure 2.
Multiple sequence alignment of all mammalian IFNE genes. Seventy-three mammalian species have available IFNE sequences, used for alignment. (A) Nucleotide sequence alignment of IFNE genes across different mammalian species. Blue = insertion, green = deletion, pink = stop codon. Protein sequence alignment of IFNE genes across different mammalian species is also shown. We identified an insertion (blue) in IFNE starting from nucleotide position 195 but not in other mammalian species, indicating that this insertion is specific to pangolins and possibly a marker to differentiate pangolins and other mammalian species. A premature stop codon or frameshift (orange) in the IFNE gene was consistently detected in both pangolin genomes. These mutations were validated by Sanger sequencing in eight Malayan pangolins. Protein sequences highlighted in orange are the frameshift mutations and premature stop codon. (B) Comparison between pangolin and human (reference) IFNE protein sequences. Pangolins have a short putative protein sequence because of a premature stop codon. Predicted functional domains and signatures in the human IFNE gene are represented by colored boxes. Yellow and pink boxes represent the predicted binding residues to IFNAR2 and IFNAR1, respectively, and which are bounded by interferons, which we obtained from a previosuly published paper (Fung et al. 2013). IFabd (SM00076) is a conserved functional domain in known interferons. The main conserved structural feature of interferons is a disulfide bond. INTERFERONAB (PR00266) is a three-element fingerprint that provides a signature for alpha, beta, and omega interferons. The elements 1 and 3 contain Cys residues involved in disulfide bond formation.