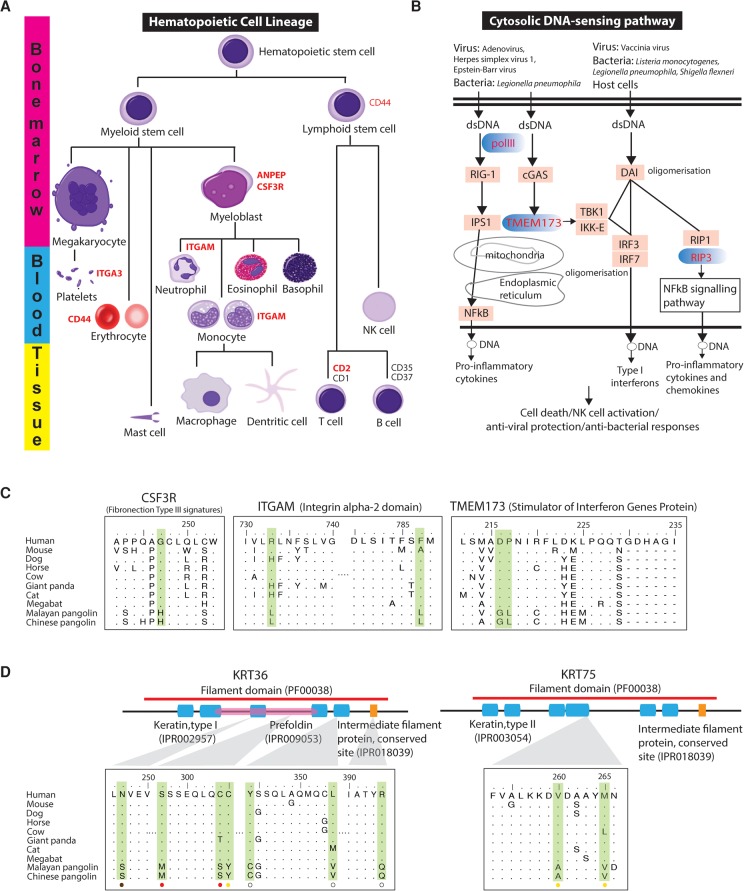
Figure 4.
Evolution of immunity-related pathways and scale-related genes in the pangolin ancestor. Genes under positive selection in the pangolin lineage in hematopoietic cell lineage (A) and cytosolic DNA-sensing pathway (B) are highlighted in red. (C) Several critical pangolin-specific mutations were identified in the functionally relevant Fibronection Type III signatures (INTERPRO ID = IPR003961) of CSF3R, the Integrin alpha-2 signatures (IPR013649) of ITGAM, and the Stimulator of Interferon Genes Protein region (INTERPRO ID = IPR029158) of TMEM173. For the CSF3R, ITGAM, and TMEM173, P-values of the detection of positive selection were 1.58 × 10−2, 3.75 × 10−2, and 2.92 × 10−5, respectively. (D) Several critical pangolin-specific amino acid changes were detected in hair-/scale-related keratin proteins, KRT36 and KRT75, which are located in functionally relevant regions that may affect protein functions. For the KRT36 and KRT75, P-values of the detection of positive selection were 2.24 × 10−2 and 1.33 × 10−5, respectively. We also examined whether African species (M. tricuspis, M. tetradactyla, and M. temminckii) have the critical pangolin-specific amino acid changes that we observed in the Asian pangolins. Circles at the bottom indicate our preliminary results: Red circle = all African species that we examined have the identical amino acid change, yellow circle = not all African species that we examined have the identical amino acid change (but they all have amino changes/deletion that likely affect protein function as predicted by PROVEAN), brown circle = all African species that we examined have the same amino acids like the human reference sequence, white circle = data not available. The alignment results are shown in Supplemental Figure S7.5.