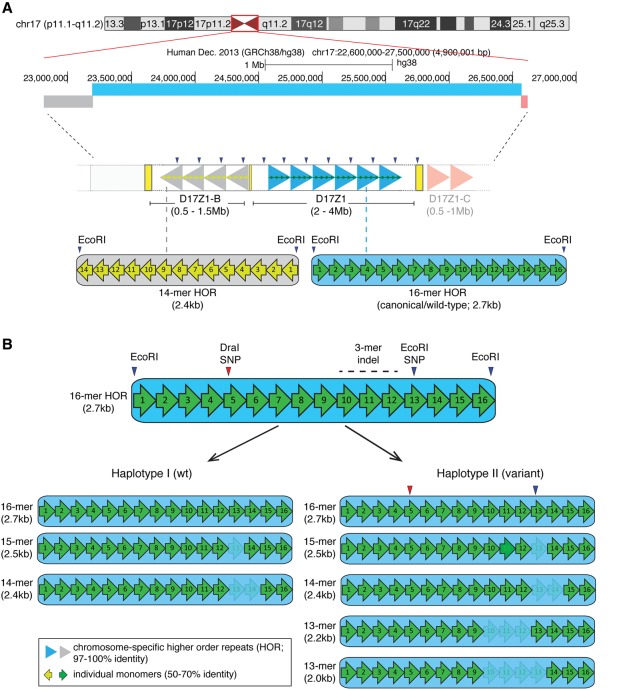
Figure 1.
Homo sapiens Chromosome 17 (HSA17) centromeric satellite organization. (A) HSA17, illustrated by the UCSC Genome Browser chromosome ideogram, has three distinct alpha satellite arrays with different monomer organizations of higher order repeats (HORs). D17Z1 (blue bar) is comprised of canonical/wild-type 16-monomer (16-mer) HORs (large blue arrowheads) that are operationally defined by EcoRI restriction sites. D17Z1-B (gray bar), located toward the short arm side of the centromere region, is based on a 14-mer HOR (large gray arrowheads) that is also defined by EcoRI sites. D17Z1-C, a third array oriented toward the long arm side of the centromere (light red bar) is also defined by a 14-mer HOR (light red arrowheads) but is less well characterized and not a focus of this manuscript. Individual monomers (green or yellow arrows) in the HORs are <70% identical to each other but are arranged nonrandomly in the same order (i.e., monomer 1 through monomer 16). The HORs are repeated hundreds to thousands of times to create highly homogenous arrays that span multiple megabases. This high degree of homogeneity has confounded standard genomic assemblies of centromere regions. (B) D17Z1 is an extremely polymorphic alpha satellite array; D17Z1-B exists exclusively as a 14-mer HOR array. In D17Z1, single and multiple monomer deletions caused by repeated rounds of unequal crossing over and/or gene conversion produce HOR variants that differ in length by an integral number of monomers. In the general population, HOR variants range from 15-mers to 12-mers, with rare 11-mers (not shown) occurring in isolated populations or individuals (Warburton and Willard 1995). Two major D17Z1 haplotypes (I, II) exist in the population and are primarily distinguished by the presence or absence of a 13-mer, created by a three-monomer deletion (3-mer indel). Additional restriction enzyme polymorphisms (DraI in monomer 5 and a second EcoRI SNP in monomer 13) are in linkage disequilibrium with the 13-mer HOR (Warburton et al. 1991). Individual monomers are denoted by numbered arrows. Shading/lightness indicates monomers that are deleted in specific HOR variants.