Figure 2.
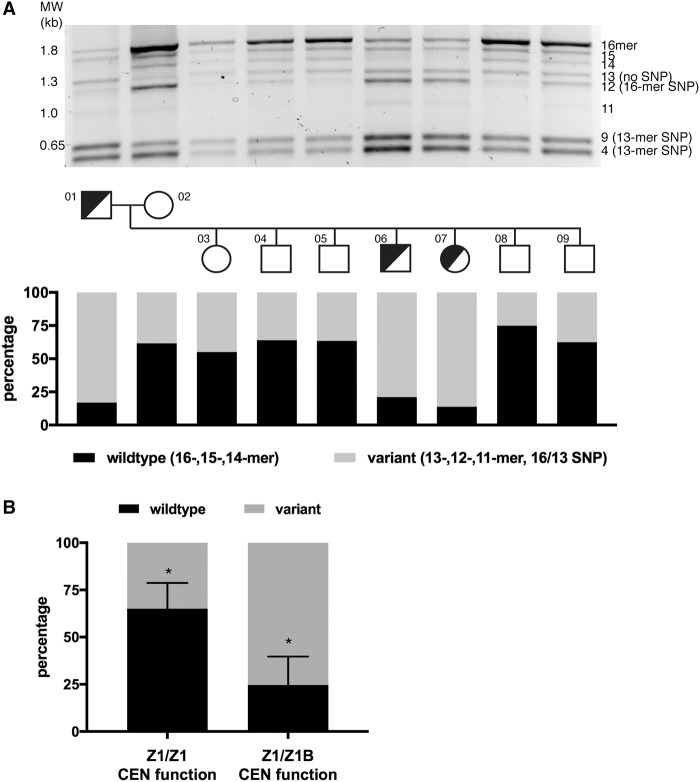
Extensive D17Z1 variation is associated with centromeric epialleles. (A) D17Z1 variation was detected using PCR followed by restriction digestion to reveal HOR size variation as well as identify HORs containing the EcoRI SNP that segregates with the 13-mer/indel HOR. D17Z1 variation within two generations of the three generation CEPH/Utah 1345 family is shown (for data on the third generation, see Supplemental Fig. S2). This family has individuals that are centromeric functional heterozygotes (half-shaded circles or squares): One homolog assembles the centromere at D17Z1 (Z1) and the other homolog assembles the centromere at D17Z1-B (Z1-B). Squares represent males; circles represent females. Each family member is numbered according to the original classification of the pedigrees (Dausset et al. 1990). Agarose gels were imaged as white bands on black background; the images were inverted for presentation purpose only. Quantitation of the amount of wild-type HORs (16-, 15-, 14-mers, Haplotype I) versus variant HORs (13-, 12-, 11-mers, and 13-mer SNP represented by 9 + 4 bands, Haplotype II) was measured. Individuals with HSA17 centromere epialleles (half-shaded squares and circles) had D17Z1 arrays with >80% variation. (B) In CEPH family 1345, the correlation between all individuals with D17Z1 variation and centromeric epialleles (D17Z1-B CEN function on one homolog only) compared to individuals lacking epialleles (D17Z1/D17Z1 CEN function) was statistically significant (asterisks indicate P < 0.001).