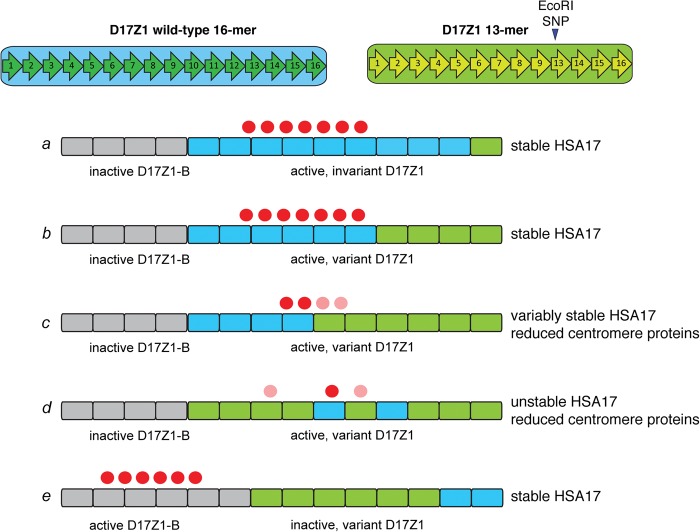
Figure 5.
Models for epiallele choice on HSA17 based on CENPB box number or D17Z1 long-range organization. Centromere assembly on HSA17 appears to occur predominantly at large D17Z1 arrays that contain wild-type (invariant, blue blocks) HORs (scenario a). When D17Z1 contains variant HORs (green blocks), centromere assembly either occurs at D17Z1-B (gray blocks, scenario e) and the HSA17 is stable, or at variant D17Z1 and the HSA17 is unstable due to reduction in CENPs (red circles). It is not clear if variant arrays cannot recruit or cannot maintain the appropriate number of CENPs, and the molecular basis for the reduction in CENP molecules is unknown. Long-range organization of D17Z1 might affect CENP recruitment and binding. Large arrays with moderate variation may provide a sufficiently sized domain of homogenous wild-type HORs for centromere assembly (scenario b). However, in the cases of HSA17s that build their centromeres on variant HSA17 arrays and are unstable, CENPA may be distributed across wild-type and variant HOR domains, the latter of which may be less efficient at CENP recruitment/maintenance (scenario c, light red circles). Moreover, irregularity in wild-type and variant HOR organization (i.e., interspersed subarrays of variant and wild-type HORs) may negatively affect centromere function or CENP recruitment (scenario d). It will be important to experimentally discriminate between these organizational scenarios, particularly on HSA17, in order to better understand the spatial relationship between centromere proteins and long-range alpha satellite organization.