Abstract
Glucocorticoids (GCs), as ligands for the glucocorticoid receptor (GR), represent one of the most effective and frequently used classes of drugs for anti-inflammatory and immunosuppressive therapy. In addition, its role in physiological and pathophysiological processes makes the GR an important research target. The past decades have yielded a wealth of insight into the physiological and pharmacological effects of GCs. Today’s era of next generation sequencing techniques is now beginning to elucidate the molecular and genomic circuits underlying GR’s cell type-specific actions. This review focuses on the concepts and insights gained from recent studies in two of the most important tissues for GC action: the liver (mediating GR’s metabolic effects) and macrophages (as the main target of anti-inflammatory GC therapy). We summarize results obtained from transgenic mouse models, molecular and genome-wide studies to illustrate GR’s complex interactions with DNA, chromatin, co-regulators and other transcription factors. Characterizing the cell type-specific transcriptional complexes assembled around GR will pave the road for the development of new anti-inflammatory and metabolic therapies in the future.
Keywords: Glucocorticoid Receptor, inflammation, metabolism, transcription, genomics, coregulators
1. Introduction
The glucocorticoid receptor (GR) belongs to the nuclear hormone receptor family of ligand-activated transcription factors. With their widespread influence on body homeostasis, metabolism, the immune system and embryonic development, nuclear receptors (NRs) are prime targets to study the nature of physiological responses in health and disease [1]. Glucocorticoids (GCs) are a class of steroid hormones that act as ligands for GR. By the middle of the last century, the anti-inflammatory and immunosuppressive potential of the GC-GR axis has gained widespread significance and relevance as powerful therapy for inflammatory diseases [2]. In normal physiology, GCs control systemic energy homeostasis and the body’s stress response through the hypothalamic-pituitary-adrenal (HPA) axis [3]. GR knockout mice do not survive beyond a few hours after birth due to respiratory failure, further demonstrating GR’s important role for survival [4]. Unfortunately, its beneficial anti-inflammatory potential used in GC therapy comes along with a wide array of detrimental side effects, many of which are linked to GR’s powerful role as a regulator of metabolic genes. Some of those metabolic side effects are shared with patients of Cushing’s disease, which present with high circulating GC levels and associated hyperglycemia, hepatic steatosis, insulin resistance and central obesity [5, 6].
At the cellular level, activated GC-bound GR translocates from the cytoplasm into the nucleus where it both positively and negatively regulates the expression of genes. GR can directly bind to glucocorticoid response elements (GREs), classically defined as inverted repeats (5′-nGnACAnnnTGTnCn-3′) in the DNA sequence of promoters and enhancers of target genes [7, 8]. GR can influence gene expression either directly or by interaction with other DNA-bound transcription factors or via composite elements (a combination of GRE and other motifs in close proximity) [9, 10]. Ligand-activated GR does not act in isolation; rather, it recruits an array of cofactors/coregulators thereby assembling large multiprotein complexes that ultimately affect gene expression.
With the increasing amount of genomic data on hand, the past years have shown that GR targets are highly regulated in a time-(circadian rhythm, signals and stimuli) and space-(cell type) dependent manner. Studies in primary macrophages and liver strongly suggest an additional layer of tissue- and context-specific regulation [11, 12]. How the same receptor can simultaneously both activate and repress target genes remains an open and equally puzzling question. Tight control over which genes are activated and which are suppressed ensures proper execution of two of GR’s signature functions: induction of glucose synthesis and suppression of inflammation. This review discusses the recent insights into gene regulation by the GR in liver and macrophages and focuses on the genomic and epigenetic mechanisms that determine the cell type- and locus-specificity of GR-mediated gene regulation.
2. Who’s talking? GR’s neighborhood in hepatocytes and macrophages
2.1 GR acts as a master regulator of glucose metabolism in liver
As a master player in maximizing circulating glucose output to maintain proper body function in times of fasting and stress, GR activates processes across different tissues which all serve the one purpose of providing substrates to the liver to fuel de novo gluconeogenesis [13]. In the liver, GR controls expression of phosphoenolpyruvate carboxykinase 1 (Pck1) and glucose-6-phosphatase (G6pc): two rate-limiting enzymes in hepatic gluconeogenesis. Functional GREs in promoters of both genes have been identified and characterized over the past 20 years and serve as the paradigm of transcriptional activation under GR control [14, 15]. By up-regulating Pck1 and G6pc expression, GR also indirectly drives hepatic fatty acid synthesis through the induction of hyperglycemia and hyperinsulinemia, two processes implicated in de novo lipogenesis [16].
Another GR target gene involved in hepatic lipid metabolism is the hairy and enhancer of split-1 (Hes1) gene. Lemke et al. demonstrated that a liver-specific knockout of GR leads to the induction of Hes1 gene expression and ameliorates steatotis in a fatty liver mouse model [17]. GR negatively interferes with Hes1 promoter activity through the recruitment of histone deacetylases. Apparently Hes1 needs to be silenced in order for GR to execute its regulatory function on genomic liver target genes [17]. This negative feedback by GR was further shown in mice with a conditional knockout of Hes1 [18]. Here, loss of hepatic Hes1 led to abnormal GR-mediated target gene regulation and a metabolic phenotype; once again demonstrating that silencing of Hes1 by GR is necessary for proper GC action [18].
Other GR-regulated target genes in liver include the circadian gene period circadian clock 1 (Per1) and the tyrosine aminotransferase (TAT) gene, which is exclusively expressed in hepatocytes. A glucocorticoid-responsive element in the Per1 promoter was shown to be indispensable for mRNA induction both in vitro and in vivo, thus tying GR to the maintenance of circadian rhythms in liver [19]. The TAT gene is another example of a metabolic enzyme under GR control [20]. In the liver, it catalyzes the rate-limiting conversion of tyrosine to 4-hydroxyphenylpyruvate, an important step in amino acid metabolism [21].
With the wealth of whole-genome Chromatin-Immunoprecipitation (ChIP-Seq) data now available, it is becoming increasingly clear that GR’s action in the liver goes way beyond glucose and lipid metabolism. Pathway and biological process analyses of GR-bound sequences (GBSs) in mouse liver [11] illustrate the myriad of direct GR binding events (Figure 1). In addition to gluconeogenesis and lipid metabolism, pathways regulated by GR are insulin signaling, PPAR signaling, bile acid metabolism and growth hormone receptor signaling, to name a few.
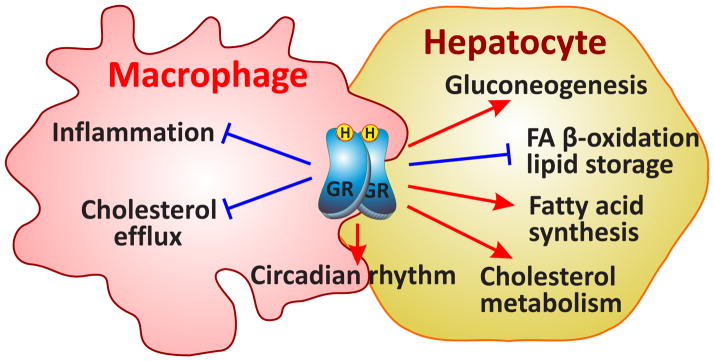
Figure 1.
Cell type-specific effects of GCs in liver and macrophages. The GR stimulates gluconeogenesis, fatty acid (FA) synthesis and cholesterol transport in liver, whereas it represses lipid storage, β-oxidation and bile acid synthesis favoring the release of glucose for a fast energy provision. Signaling pathways affected by GR are Peroxisome Proliferator Activated Receptor (PPAR) and growth hormone receptor (GhR) signaling. The main function of GR in macrophages is its repressive actions on inflammation via inhibition of cytokine production, suppression of interleukin 6 (Il6) and lipopolysaccharide (LPS) signaling. Circadian rhythm is influenced by glucocorticoids in both tissues. → activation; ⊣ repression
2.1.1 Endowing competence for hepatocyte cell fate: Neighboring motifs in liver
By motif enrichment analysis of ChIP-Seq data sets for GR in mouse liver, transcription factor footprints for CCAAT/enhancer binding protein (c/EBP), hepatic nuclear factor 4 (Hnf4), Onecut-1 (Oc1) and the Forkhead factor family (Fox) have been identified as significantly enriched together with GREs (Figure 2) [7, 11, 22]. c/EBP was previously shown to be indispensable for GR access and binding of the DNA in mouse livers [11]. Indeed, disruption of c/EBP binding to the DNA itself led to loss of chromatin accessibility and interfered with GR recruitment and DNA binding [11]. Interestingly, Hnf4, Oc1 as well as members of the Forkhead factor family all have crucial roles in liver development, thus supporting their role as lineage-determining factors. Whereas Hnf4 is essential for mammalian hepatocyte differentiation [23] and maintenance of hepatic gene expression [24], Oc1 (or Hnf6) controls early hepatoblast migration [25] and coordinates time-specific gene expression during liver development [26]. Intriguingly, FoxA2 (also called Hnf3) is suggested to be involved in the establishment of competence by opening up closed chromatin in liver-specific genes. Gualdi et al. demonstrated that hepatic differentiation from the gut endoderm is characterized by HNF3 occupancy at the albumin enhancer, which keeps the enhancer transcriptionally silent but open for additional factor binding and gene activation [27]. Moreover, Foxa1 and Foxa2 knockout mice fail to develop a liver bud, further corroborating their crucial role for the onset of hepatogenesis [28]. While the ChIP-Seq approach can determine the presence of these factors, it is lacking the resolution required for the accurate mapping of binding sites. ChIP-exo (ChIP-Seq coupled to exonuclease digestion) is a new technique that overcomes this limitation and allows for more precise determination of binding sites at almost single base pair resolution. Starick et al. made use of ChIP-exo to identify Oc1 and FoxA as the closest TF footprints to GREs in different cell lines [8]. Moreover, Lim et al. identified the same two motifs by ChIP-exo in mouse liver, supporting their association with GREs in the hepatic cistrome [7].
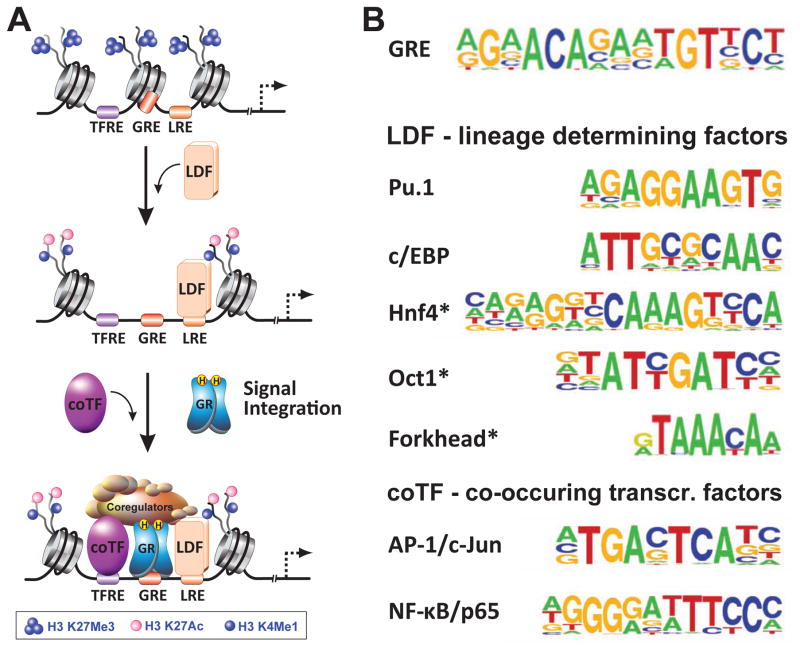
Figure 2.
Mechanism of cell type-specific gene regulation by GR. (A) Lineage-determining factors (LDFs) are transcription factors that recognize their DNA binding motif (LRE) within compacted chromatin and recruit chromatin remodeling machinery to “open” it and to reposition nucleosomes. Subsequent recruitment of histone demethylases/methylases and histone acetyltransferases establishes a cell type-specific enhancer landscape. “Open” chromatin is marked by increased DNA accessibility and specific histone marks, one being acetylation at H3K27, replacing a methylation mark at this residue (H3K27me3) associated with repressed chromatin. Enhancers are marked by a monomethylation mark at H3K4, and active enhancers harbor the double label H3K4me1/H3K27ac. Once the enhancer landscape of a cell is specified, signal-dependent transcription factors such as GR are able to access their response elements (GREs). Together with co-occuring transcription factors (coTF), they regulate target genes in a locus-specific manner. The combinatorial diversity of GREs and TFREs (response elements of co-occuring transcription factors) is the hallmark of gene-specific outcomes of GR signaling. (B) Transcription factor binding motifs identified as co-occuring with GREs in ChIP-Seq experiments in liver and macrophages can be classified as LREs or TFREs.
Besides their requirement in liver development, c/EBP [29], Hnf4 [30], FoxA2 [31] and FoxO1 [32] all enable recruitment of GR to liver-specific promoters and enhancers, such as the Pck1 locus [33].
2.2 GR signaling in macrophage inflammation
For over 60 years, patients have greatly benefited from the potent anti-inflammatory effects of GCs, making them the most widely prescribed drugs in anti-inflammatory therapy. GCs bind GR in a variety of immune cells and stimulate anti-inflammatory actions that differ depending on the stimulus and disease model. In this context, macrophages are at the front line of the innate immune response and the major targets in the resolution of inflammation in sepsis and contact allergies [34, 35]. Importantly, cytokine and chemokine production is efficiently suppressed by GR in a transcription-dependent manner (Figure 1) [36].
In macrophages, GR activates expression of both anti-inflammatory genes such as glucocorticoid-induced leucine zipper (Tsc22d3/Gilz), dual specificity phosphatase/MAP kinase phosphatase 1 (Dusp1) or krueppel-like factors 2 and 9 (Klf2, Klf9), and circadian genes such as period circadian clock 1 (Per1). However, the crucial anti-inflammatory function of ligand-bound GR in macrophages is the repression of inflammatory genes. This latter class of GR targets encodes classic inflammatory cytokines such as interleukin 6 (Il6), interleukin 1 alpha/beta (Il1a/b), chemokines such as chemokine (C-C motif) ligand 2 or 5 (Ccl2, Ccl5) or chemokine (C-X-C motif) ligand 10 (Cxcl10) and other inflammatory mediators such as inducible nitric oxide synthase 2 (Nos2), the matrix metalloproteases 12 and 13 (Mmp12/13) and tumor necrosis factor (Tnf) [12, 34].
2.2.1 Towards a myeloid lineage: Role of priming factors in macrophages
Genome-wide analyses of GR binding events by ChIP-Seq confirmed GR recruitment to enhancers regulating inflammatory genes, but did not yield the anticipated insight into defined regulatory sequences discriminating between activating and repressing regulatory elements [12]. A closer look at co-occurring transcription factor binding sites identified Spleen focus forming virus (SFFV) Proviral Integration oncogene (Spi1 also known as Pu.1), c/EBP, jun proto-oncogene (cJun) and v-rel ReticuloEndotheLiosis viral oncogene homolog A (RelA also known as p65) as possible binding partners at enhancers of both activated and repressed genes in macrophages (Figure 2) [7, 12]. Interestingly, Pu.1 and c/EBP are essential for development of the myeloid lineage [37, 38]. Loss of Pu.1 in hematopoietic stem cells completely blocks their self-renewing capabilities. Furthermore, Pu.1 remains expressed after lineage commitment of hematopoietic stem cells towards lymphoid and myeloid lineages and is required for myeloid maturation, but not for proliferation or survival [38]. Conversely, loss of c/EBPα increases the proliferation and self-renewing capabilities of hematopoietic stem cells, and blocks the M-CSF-dependent differentiation towards the myeloid lineage [39, 40]. Thus, both Pu.1 and c/EBP are required to balance hematopoietic stem cell proliferation versus differentiation, and have combinatorial functions in “priming” the myeloid lineage and establishing the monocyte-specific enhancer landscape [37, 41]. Additionally, other c/EBP isoforms, e.g. c/EBPβ and c/EBPε are required for myeloid differentiation [42] and inflammatory responses in macrophages [37]. In addition to their function in setting the myeloid enhancer landscape, a direct interaction of GR with c/EBPα in human primary cells was shown [43].
Furthermore, direct protein-protein interactions of GR with the NF-κB subunit p65 in Hela cells under overexpression conditions [44] or in vitro [45] and the AP-1 subunit c-Jun in vitro [46] was demonstrated. Additionally, in 2011 Biddie and colleagues showed that AP-1 (c-Jun/c-Fos heterodimer) is necessary for GR recruitment to a subset of AP-1/GR composite elements in 3134 cells [47]. Similarly, upon TNFalpha stimulation, Hela cells gain a subset of GREs associated with pro-inflammatory genes dependent on the presence of NF-κB/p65 by co-occupying those regulatory elements [48]. This strengthens the argument that GR-mediated gene regulation depends on the simultaneous action of AP-1/c-Jun and NF-κB/p65. Interestingly, a closer look at the repressed GR cistrome revealed the Interferon Regulatory Factor 3 (IRF3) as an additional binding partner that might be involved in GR repression in macrophages [12]. Interestingly, in 2005 Ogawa and colleagues proposed a mechanism by which GR interferes with the lipopolysaccharide (LPS)-induced p65/IRF3 interaction at a subset of shared enhancers [45].
Clearly, although a deeper insight into GR-mediated pro- and anti-inflammatory gene regulation has been gained in recent years, much remains to be learned regarding the mechanisms underlying selective gene repression and activation by GC-bound GR in macrophages.
3. GR and its coregulators
Coregulators typically act via three distinct mechanisms to ultimately link upstream cellular signaling events from the environment into functional genomic responses: by remodeling chromatin in an ATP-dependent manner, by covalently modifying histones or other components of transcription complexes (or recruiting secondary cofactors with such activities), or by directly facilitating the assembly of basic transcriptional machinery. Interestingly, growing evidence suggests that coregulators function in a target gene-specific manner. Thus, although the exact mechanisms that dictate specificity of cofactor utilization by GR remain poorly defined, these proteins potentially represent attractive candidates that can be manipulated in order to bias GR towards specific regulatory programs. In the following section, we discuss recent studies pointing to less typical and perhaps unexpected coregulator activities in regards to GR-driven gene expression.
3.1 Nuclear Receptor Coactivators (NCOAs) of the p160 family
The most well characterized nuclear receptor coregulators are adaptor proteins of the p160 family, NCOA1/SRC1, NCOA2/TIF2/GRIP1/SRC2, and NCOA3/pCIP/ACTR/AIB1/SRC3. The importance for these proteins in diverse biological processes including metabolism, circadian rhythm, immunity and reproduction as revealed by gene ablation studies in mice has been extensively reviewed elsewhere [49–51].
All p160s are recruited by the AF2 regions of agonist-activated NRs via one of three LXXLL motifs (NR boxes; GR interacts preferentially with NR box-3) in their NR interaction domain (NID) [52, 53]. Acting as binding platforms for numerous secondary chromatin modifiers such as CBP1/p300, PRMT1, CARM1, the Baf57 subunit of the SWI/SNF complex and other cofactors, p160 family members ultimately potentiate the activation of associated target genes. Because much of the molecular characterization of the p160 coactivator functions as they relate specifically to GR has been performed using in vitro and overexpression systems, the involvement of individual p160s in driving transcription of GC-responsive genes and, hence, their contribution to specific GR-driven biological pathways in vivo, remains relatively unclear. Future genome-wide analyses of GR transcription programs using different cell types lacking p160s in question will be instrumental in linking a wealth of structural and biochemical data on p160 function to GC-regulated physiological processes.
Notably, with respect to GR, one p160 family member – NCOA2 or GR-interacting protein 1 (GRIP1) – stands out. In addition to its coactivator properties, it is the only p160 family member to have emerged as a key GR corepressor at GR, AP-1 and NF-κB co-occupied sites [54, 55]. GRIP1 was found to be recruited to such sites in conjunction with liganded GR and potentiate repression of associated genes, e.g., Tnf, Ccl2, Ccl3, Ccl4, Il1a, Il1b and many others [12]. In fact, at the transcriptomic level, almost half of genes induced by inflammatory triggers such as LPS and repressed by liganded GR in murine bone marrow-derived macrophages (BMMϕ) lost their GC sensitivity in GRIP1-deficient BMMϕ [56].
The mechanistic basis of GR:GRIP1-mediated repression is not fully understood, however this activity appears to require a unique domain of GRIP1 not conserved across the p160 family [54]. Moreover, many genes susceptible to repression by GR:GRIP1 were found to be targeted at the post-initiation step. Historically, signal-dependent transcription factors and, their coregulators were thought to affect chromatin and components of basal transcriptional machinery ultimately facilitating or precluding RNA Polymerase II recruitment and ultimately transcription initiation [57]. Indeed, genes such as Il1b and Il1a are repressed by GR:GRIP1 via inhibition of Pol II recruitment and transcription initiation (Figure 3A, left) [58]. In recent years, however, an alternative picture has emerged whereby early transcription elongation represents a critical rate-limiting step for up to 40% of regulated genes [59, 60]. At such genes, Pol II initiates transcription prior to stimulation but pauses ~50 nucleotides from the TSS bound by the Negative Elongation Factor (NELF) [59–62]. Upon exposure to relevant stimulus, positive transcription elongation factor P-TEFb, composed of cyclin T1 and CDK9, phosphorylates Pol II as well as NELF triggering its dissociation and the release of Pol II into productive elongation (Figure 3A, right). This level of regulation is very common among genes encoding inflammatory cytokines such as TNF, CCL3 and CCL2 [63], all of which are also targets for GR:GRIP1-mediated repression. Furthermore, it was shown in primary macrophages that at those genes, it was signal-dependent pause release that was attenuated by GR:GRIP1. Specifically, GR:GRIP1 complex inhibited P-TEFb recruitment and triggered NELF accumulation at the TSS (Figure 3A, right) [58, 63]. Consistent with a critical role of NELF in GR-mediated repression specifically of such ‘elongation-controlled’ genes, their response to Dex was blunted in NELF-deficient macrophages [58]. Importantly, GR-imposed control of pause release required GRIP1 as the attenuation of P-TEFb recruitment and the retention of NELF were both abrogated by GRIP1 deletion, thus rendering these pro-inflammatory genes GC-resistant [58]. This unexpected finding points to previously unappreciated function that GRIP1 and perhaps other coregulators may play in controlling gene expression at post-initiation steps.
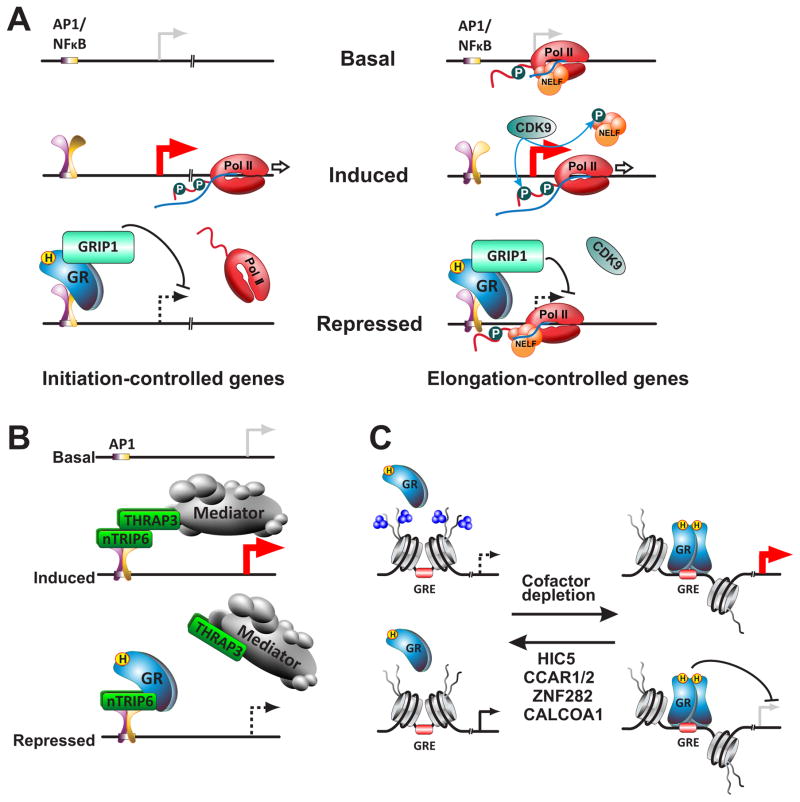
Figure 3.
Mechanisms of GR coregulator function A. GR:GRIP1 repress initiation- and elongation-controlled genes by distinct mechanisms. Initiation-controlled genes: GR:GRIP1 complex blocks the recruitment of Pol II to the TSS, the rate-limiting step for gene activation. Elongation-controlled genes: GR:GRIP1 complex blunts CDK9 recruitment, and promotes accumulation of NELF at the TSS thus blocking phosphorylation of Serine 2 of the Pol II C-terminal domain and entry into productive elongation. B. nTRIP6-facilitated repression by GR. In the presence of activating signals such as TPA, nTRIP6 acts as a scaffold for the recruitment of THRAP3 and the Mediator complex to Fos-containing AP-1 dimers thus potentiating activation of associated target genes. Upon GC exposure, GR binds to nTRIP6 and precludes its interaction with THRAP3 and Mediator, thereby attenuating target gene transcription. C. Coregulators dictate GR binding site selection. In the presence of the coregulators HIC5, ZNF282, CCAR1/2, or CALCOA1, a subset of potential GREs are associated with inaccessible chromatin and the repressive histone mark, H3K27me3. Upon coregulator depletion, chromatin accessibility increases and GR can bind the open binding sites and regulate transcription of associated genes.
3.2 TRIP6
TRIP6 is a member of the zyxin family with roles in focal adhesion, which contains LIM domains involved in protein:protein interactions. The nuclear isoform of TRIP6, nTRIP6, is thought to serve as an adaptor between GR and Fos-containing AP-1 dimers, facilitating hormone-dependent recruitment of GR to AP1 complexes in the context of “tethering” sites [64–66]. In the absence of hormone, nTRIP6 acted as an AP-1 coactivator by recruiting THRAP3 and MED1 subunits of the mediator complex (Figure 3B). Indeed, introduction of peptides that disrupt nTRIP6 dimerization and interaction with the mediator resulted in blunted recruitment of THRAP3 and MED1 to an AP-1-driven Mmp13 promoter in response to phorbol ester (e.g., TPA) and transcriptional activation [65]. In the presence of GC, however, GR bound nTRIP6 thereby precluding the nTRIP6:THRAP3 and nTRIP6:MED1 interactions and attenuating gene activation in response to TPA (Figure 3B). The repressive effect of liganded GR was abrogated when nTRIP6 was depleted, although the extent to which gene repression by GR vs. gene activation by AP-1 was affected by TRIP6 knockdown is difficult to determine [67]. In addition, given that much of this work has been performed in cell lines with overexpression constructs, it will be essential to extend these studies to gene silencing with endogenous systems in inflammatory cell types to gauge their physiological relevance.
3.3. Hydrogen Peroxide-Inducible Clone 5 (HIC5)
HIC5/TGFB1l1 is, like TRIP6 or Paxillin, a LIM domain protein involved in focal adhesion, that also serves as a coregulator for multiple NRs. HIC5 binds the τ2 hinge domain of human GR [68] and can function to either potentiate or attenuate GR actions. The unique feature of HIC5, however, was revealed through genome-wide studies following HIC5 siRNA-mediated depletion in human osteosarcoma cells overexpressing GR, U2OS-GR [69]. In this study, about 40% of Dex-regulated genes were affected by HIC5 depletion, with the vast majority of those becoming less Dex-responsive (i.e., genes activated by Dex were less activated, and genes repressed by Dex were less repressed) in HIC5-deficient cells relative to wild-type. This is in agreement with conventional actions of HIC5 as a GR coregulator as ChIP-Seq revealed GBS near many of these candidate Dex-activated genes and ChIP-qPCR analysis showed that HIC5 depletion resulted in decreased recruitment of MED1 and Pol II to their TSS. The unexpected finding, however, was that a significant number of genes acquired Dex responsiveness only following HIC5 knockdown. Indeed, those so-called “blocked” genes displayed sites of Dex-induced GR occupancy as well as p300/CBP, MED1 and Pol II recruitment to the TSS that gained positive Dex regulation upon depletion of HIC5, which also correlated with greatly increased chromatin opening as assessed by FAIRE-qPCR (Figure 3C). These results illustrate a non-traditional role for HIC5 as a cofactor in that by selectively promoting chromatin remodeling, it dictates binding site selection by GR. The described scenario challenges an established model, according to which cofactors are passively recruited to chromatin by DNA-bound regulators. However, its physiological role is so far unknown and presents a fascinating topic for future studies.
HIC5 interacts with multiple steroid receptors; interestingly, however, despite binding GRα (the major GR isoform), GRγ and ERα similarly, its depletion affected the greatest number of GRα-regulated genes [70]. Still, “blocked” genes, whose increased chromatin accessibility and hormonal regulation was unveiled through HIC5 knockdown, were found for all steroid receptors analyzed. These studies therefore point to a previously unappreciated active function of coregulators in dictating the specificity of transcription factor actions. Indeed, for some factors for which genome-wide data are available, there appear to be vastly more potential binding sites present than is actually being utilized (e.g., AP-1) [71], and what determines tissue-specific binding remains poorly understood. A role for coregulators in limiting chromatin accessibility to GR or other factors represents a compelling mechanism that would enable binding site selection to operate in a cell-specific manner and, thus, an exciting new avenue for future studies.
4. Shaping cell type specification
4.1 Role of the epigenetic landscape in GR gene regulation
The epigenetic landscape of a cell is a major contributor to cell type-specific responses upon stimulation. GCs have long been known to elicit tissue-specific effects. Whereas the physiological function of GCs in providing glucose for energy production is mainly mediated by hepatocytes, their most relevant function in clinical practice lies in the anti-inflammatory actions in immune cells.
Target genes of the ligand-bound GR form two groups according to their cell type-specific behavior. The first group is composed of genes, which are regulated by GR independently of cellular context like the maintenance of basal cellular processes or circadian effects, e.g. Per1 or Klf9 [11, 12, 72]. The second group includes genes with their expression changing in some but not in other tissues upon GC treatment, e.g. the metabolic tissue-specific genes Pck1 [73] and G6pc [74, 75] or the inflammatory cytokines Il6 and Il1a, which are repressed by GR in macrophages [12], but not in hepatocytes.
ChIP-Seq experiments on GR have revealed that GR binds different enhancers depending on the cell type. It was for example shown that GR binds the enhancer and promoter regions of Pck1 and G6pc in liver [11], but not in macrophages treated with LPS. On the other hand, GR binds the enhancers of Il6 and Il1a in macrophages after LPS treatment [12] but not in hepatocytes (Figure 4). A closer look at the epigenetic signature revealed that both the Pck1 and G6pc enhancer are acetylated at H3K27 and monomethylated at H3K4 in hepatocytes [76] but not in activated macrophages [57]. The same applies to the enhancers of Il6 and Il1a. Indeed, up to a third of GR-bound sites in liver and macrophages overlap with H3K4me1 and H3K27Ac chromatin that is modified in the absence of GR and its ligand. (Figure 4) [11, 12, 57, 76]. This indicates the requirement of active enhancers [77] and open chromatin for GR binding in both liver [11] and macrophages. Similarly, results from John and colleagues demonstrated that GR requires “open” chromatin for binding in 3134 and AtT-20 cells [78]. Therefore, cell type-specific actions of GR appear to depend on the epigenetic landscape and DNA accessibility rather than on chromatin remodeling and enhancer priming by GR itself.
Figure 4.
Chromatin accessibility mediates cell type-specific actions of GR. ChIP-Seq tracks for GR, H3K4me3, H3K4me1 and H3K27ac in liver and inflammatory macrophages. (A) For Per1, activated by GR in both tissues, the GR peaks lie in close proximity to H3K4 monomethylated chromatin – a histone mark associated with enhancers. This enhancer is active in hepatocytes (overlapping H3K27ac) or poised in macrophages (missing H3K27ac). (B) In hepatocytes but not macrophages, GR binds close to the Pck1 promoter. The surrounding chromatin harbors the H3K4me1/H3K27ac double mark for active enhancers as well as a H3K4 trimethyation mark, indicating the proximity to the Pck1 promoter. Macrophage chromatin at this locus contains neither enhancer/promoter-specific histone marks nor a GBS. (C) The Il6 enhancer contains a GBS in macrophages, which overlaps with H3K4me1/H3K27ac chromatin, indicating an active enhancer. In hepatocytes this chromatin is not accessible for GR binding and harbors none of these marks.
Motif prediction analysis in GR ChIP-Seq experiments revealed that Pu.1 and c/EBP are among the most abundant motifs co-localizing with GREs in macrophages (Figure 2) [12] whereas in liver, GREs co-occur with c/EBP, Hnf4, Oc1 and Forkhead motifs (Figure 2) [11]. Pu.1 and c/EBP in macrophages as well as Hnf4, Oc1, c/EBP and Forkhead proteins in hepatocytes, respectively, are required for the development of those lineages, thus defining them as lineage-determining factors [23–28, 37–41]. Closer analysis of the combinatorial requirements of transcription factors for lineage specification and integration of cell type-specific signal responses in macrophages was consistent with a hierarchical mechanism underlying transcription factor networks [79, 80]. First, lineage-determining factors establish the basic cell type-dependent accessibility of the DNA, followed by integration of environmental and, lastly, temporal stimuli (Figure 2) [81, 82].
In conclusion, the cell type-specificity of the GR transcriptome is a consequence of lineage-specific enhancer accessibility and activity. The diversity in GR responses is determined by the cooperativity of cell type-, environment- or signal-dependent transcription factors. Studying how the temporally constrained or continuous actions of these factors influence enhancer remodeling, GR occupancy and the recruitment of coregulators will yield important insights in years to come.
4.2 Coregulator specificity towards certain physiological pathways
Gene regulation by GCs is pleiotropic and affects diverse physiological pathways. Coregulators have long been thought to impart a level of specificity to GR transcriptional regulation, albeit it was attributed primarily to tissue-specific coregulator expression. More recently, a combination of genome-wide transcriptomics combined with systematic depletion of individual coregulators suggested that distinct GC-driven physiological pathways within the same cell type display differential cofactor requirements. Indeed, siRNA-mediated depletion of CCAR1, and not other 9 coregulators analyzed in 3T3-L1 murine pre-adipocytes, greatly attenuated Dex induction of adipogenic genes but not of select anti-inflammatory genes [83]. Consistently, knockdown of CCAR1 by shRNA in mouse mesenchymal stem cells (MSCs) blunted adipocyte differentiation in response to the adipogenic cocktail Dex/insulin/ibmx/rosiglitazone. To tease out the effects of CCAR1 resulting specifically from its cooperativity with GR, Dex only treatment was used to identify major regulatory GBS in the PPARγ gene, a master regulator of adipocyte differentiation. ChIP- and FAIRE-qPCR analysis in MSCs revealed that GR and CCAR1 were recruited to these GBS in conjunction with chromatin opening at the PPARγ promoter – all of which was abrogated by CCAR1 depletion. In this regard, it would be informative to evaluate the phenotype of adipocyte-specific CCAR1 knockout mice in vivo.
A similar genome-wide analysis of the Dex-responsive transcriptome was performed in A549 cells following siRNA-mediated depletion of either CCAR1, CCAR2, CALCOCO1 or ZNF282 [84]. Not surprisingly, the vast majority of genes affected by coregulator depletion displayed decreased induction by Dex, with ZNF282 knockdown affecting the greatest number of genes (~3000, 3× more than the others). Of these genes, 1/3 were factor-specific with distinct polarity of regulation: CCAR1- and 2-unique genes induced by Dex were attenuated by their depletion – supporting the role of CCAR1 and 2 as GR coactivators; conversely, CoCoA- and ZNF282-unique genes induced by Dex were superactivated by their knockdown suggesting that CoCoA- and ZNF282 inhibit GR activity. Similar to the HIC5 studies, this experiment revealed “blocked” genes that became Dex-regulated only after cofactor depletion concomitantly with the loss of the repressive histone mark H3K27me3, suggesting a role for these coregulators in limiting chromatin accessibility to GR (Figure 3C). Interestingly, “blocked” genes were virtually all coregulator-specific and the majority (50–75%) gained positive regulation by Dex. Given the therapeutic utility of GCs, Ingenuity Pathway Analysis (IPA) was used to assess the impact of different cofactors on pathways that contribute to the anti-inflammatory effects of Dex. IPA of the acute phase response supported a significant role of ZNF282 and CoCoA in GR-mediated repression of the NF-κB pathway as well as in limiting glucocorticoid induction of STAT3, and of ZNF282 in glucocorticoid repression of c-Jun and the JNK pathway. In contrast, all coregulators but CCAR2 appeared to have similar effects on interferon signaling by limiting the Dex-induced expression of JAK1 and STAT1. CCAR2 was the exception, as IPA suggested that it potentiated Dex-mediated inhibition of interferon receptor expression. This limited analysis already reveals the ability of GR coregulators to preferentially participate in specific physiological pathways. It will be interesting to see this type of characterization extended to other cell types and tissues, such as those with key role in inflammation and metabolism given the dire need for more selective GC-like molecules in clinical practice.
5. Future directions
Recent genome-wide studies have provided unprecedented insights into the molecular and genomic mechanisms of GR action. Despite the increasing identification of binding sites, target genes and coregulators, however, we are only beginning to decipher GR’s distinctive actions across the different cell types.
High-resolution mapping of GBS with new next generation sequencing techniques such as ChIP-exo as well as advanced bioinformatic analyses will yield additional clues on the genomic information that specifies enhancers. Furthermore, the decoding of locus-specific GR/cofactor complexes by high-resolution mass spectrometry will deepen our understanding of the combinatorial complexity at GR-regulated enhancers and the role of signal integration via post-translational modifications. An emerging role of long non-coding RNAs in gene regulation provides an additional layer of complexity as does the 3-dimensional architecture of the nucleus; two aspects which so far have been insufficiently studied in the context of GR action. Furthermore, studies in large non-synchronized cell populations and heterogeneous tissues merely provide a cumulative snapshot of the underlying molecular events. Here, differences between individual cells based on cell cycle, environmental signals and positional information provided by signaling gradients or tissue architecture are neglected. With the continuous advancement of both next generation sequencing and high resolution imaging techniques towards higher sensitivity and smaller scales, we can anticipate that single cell analyses, in particular in-situ approaches, will further deepen our understanding of GC signaling in the near future.
Acknowledgments
We apologize to all authors whose work could not be cited due to the limited scope of this review. We thank Dr. Y. Chinenov for preparation of the figures. This work was supported by grants UH 275/1-1 (DFG) to FG, MCH and NHU; 638573 SILENCE (ERC-2014-StG) to NHU; a Daimler Benz Scholarship to FG; the NIH/NIDDK (R01 DK099087), The Department of Defense (PR130049) and The Rheumatology Research Foundation to IR and the NIH/NIAMS T32 AR007281 to DAR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240(4854):889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vegiopoulos A, Herzig S. Glucocorticoids, metabolism and metabolic diseases. Mol Cell Endocrinol. 2007;275(1–2):43–61. doi: 10.1016/j.mce.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Cole TJ, et al. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9(13):1608–21. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- 5.Schacke H, Docke WD, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther. 2002;96(1):23–43. doi: 10.1016/s0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 6.Shibli-Rahhal A, Van Beek M, Schlechte JA. Cushing’s syndrome. Clin Dermatol. 2006;24(4):260–5. doi: 10.1016/j.clindermatol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Lim HW, et al. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015;25(6):836–44. doi: 10.1101/gr.188581.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starick SR, et al. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 2015;25(6):825–35. doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10(5):365–76. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 10.Langlais D, et al. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47(1):38–49. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Grontved L, et al. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32(11):1568–83. doi: 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlenhaut NH, et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell. 2013;49(1):158–71. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel R, Williams-Dautovich J, Cummins CL. Minireview: new molecular mediators of glucocorticoid receptor activity in metabolic tissues. Mol Endocrinol. 2014;28(7):999–1011. doi: 10.1210/me.2014-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai E, et al. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1990;10(9):4712–9. doi: 10.1128/mcb.10.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Kooi BT, et al. The glucose-6-phosphatase catalytic subunit gene promoter contains both positive and negative glucocorticoid response elements. Mol Endocrinol. 2005;19(12):3001–22. doi: 10.1210/me.2004-0497. [DOI] [PubMed] [Google Scholar]
- 16.Sun Z, Lazar MA. Dissociating fatty liver and diabetes. Trends Endocrinol Metab. 2013;24(1):4–12. doi: 10.1016/j.tem.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemke U, et al. The glucocorticoid receptor controls hepatic dyslipidemia through Hes1. Cell Metab. 2008;8(3):212–23. doi: 10.1016/j.cmet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Revollo JR, et al. HES1 is a master regulator of glucocorticoid receptor-dependent gene expression. Sci Signal. 2013;6(304):ra103. doi: 10.1126/scisignal.2004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto T, et al. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem. 2005;280(51):42036–43. doi: 10.1074/jbc.M509600200. [DOI] [PubMed] [Google Scholar]
- 20.Jantzen HM, et al. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987;49(1):29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- 21.Natt E, et al. Point mutations in the tyrosine aminotransferase gene in tyrosinemia type II. doi: 10.1073/pnas.89.19.9297. (0027-8424 (Print)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phuc Le P, et al. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1(2):e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14(4):464–74. [PMC free article] [PubMed] [Google Scholar]
- 24.Hayhurst GP, et al. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21(4):1393–403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margagliotti S, et al. The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev Biol. 2007;311(2):579–89. doi: 10.1016/j.ydbio.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Beaudry JB, et al. Threshold levels of hepatocyte nuclear factor 6 (HNF-6) acting in synergy with HNF-4 and PGC-1alpha are required for time-specific gene expression during liver development. Mol Cell Biol. 2006;26(16):6037–46. doi: 10.1128/MCB.02445-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gualdi R, et al. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10(13):1670–82. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 28.Lee CS, et al. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435(7044):944–7. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 29.Park EA, et al. Relative roles of CCAAT/enhancer-binding protein beta and cAMP regulatory element-binding protein in controlling transcription of the gene for phosphoenolpyruvate carboxykinase (GTP) J Biol Chem. 1993;268(1):613–9. [PubMed] [Google Scholar]
- 30.Hall RK, Sladek FM, Granner DK. The orphan receptors COUP-TF and HNF-4 serve as accessory factors required for induction of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Proc Natl Acad Sci U S A. 1995;92(2):412–6. doi: 10.1073/pnas.92.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JC, et al. Hepatic nuclear factor 3 is an accessory factor required for the stimulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Mol Endocrinol. 1996;10(7):794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 32.Hall RK, et al. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J Biol Chem. 2000;275(39):30169–75. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- 33.Chakravarty K, et al. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol Biol. 2005;40(3):129–54. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- 34.Kleiman A, et al. Glucocorticoid receptor dimerization is required for survival in septic shock via suppression of interleukin-1 in macrophages. FASEB J. 2012;26(2):722–9. doi: 10.1096/fj.11-192112. [DOI] [PubMed] [Google Scholar]
- 35.Tuckermann JP, et al. Macrophages and neutrophils are the targets for immune suppression by glucocorticoids in contact allergy. J Clin Invest. 2007;117(5):1381–90. doi: 10.1172/JCI28034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuckermann JP, et al. Molecular mechanisms of glucocorticoids in the control of inflammation and lymphocyte apoptosis. Crit Rev Clin Lab Sci. 2005;42(1):71–104. doi: 10.1080/10408360590888983. [DOI] [PubMed] [Google Scholar]
- 37.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106(5):1590–600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heath V, et al. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood. 2004;104(6):1639–47. doi: 10.1182/blood-2003-11-3963. [DOI] [PubMed] [Google Scholar]
- 40.Hohaus S, et al. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol. 1995;15(10):5830–45. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin F, et al. PU.1 and C/EBP(alpha) synergistically program distinct response to NF-kappaB activation through establishing monocyte specific enhancers. Proc Natl Acad Sci U S A. 2011;108(13):5290–5. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akagi T, et al. In vivo deficiency of both C/EBPbeta and C/EBPepsilon results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5(11):e15419. doi: 10.1371/journal.pone.0015419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudiger JJ, et al. Interaction of C/EBPalpha and the glucocorticoid receptor in vivo and in nontransformed human cells. Faseb j. 2002;16(2):177–84. doi: 10.1096/fj.01-0226com. [DOI] [PubMed] [Google Scholar]
- 44.Ray A, Prefontaine KE. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1994;91(2):752–6. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa S, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122(5):707–21. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond MI, et al. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249(4974):1266–72. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 47.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43(1):145–55. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao NA, et al. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011;21(9):1404–16. doi: 10.1101/gr.118042.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dasgupta S, Lonard DM, O’Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279–92. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dasgupta S, O’Malley BW. Transcriptional coregulators: emerging roles of SRC family of coactivators in disease pathology. J Mol Endocrinol. 2014;53(2):R47–59. doi: 10.1530/JME-14-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rollins DA, Coppo M, Rogatsky I. Minireview: nuclear receptor coregulators of the p160 family: insights into inflammation and metabolism. Mol Endocrinol. 2015;29(4):502–17. doi: 10.1210/me.2015-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12(21):3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heery DM, et al. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 54.Rogatsky I, et al. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci U S A. 2002;99(26):16701–6. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. Embo j. 2001;20(21):6071–83. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chinenov Y, et al. Glucocorticoid receptor coordinates transcription factor-dominated regulatory network in macrophages. BMC Genomics. 2014;15:656. doi: 10.1186/1471-2164-15-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1–2):157–71. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 58.Gupte R, et al. Glucocorticoid receptor represses proinflammatory genes at distinct steps of the transcription cycle. Proc Natl Acad Sci U S A. 2013;110(36):14616–21. doi: 10.1073/pnas.1309898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39(12):1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39(12):1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–77. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 63.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106(43):18207–12. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diefenbacher M, et al. Restriction to Fos family members of Trip6-dependent coactivation and glucocorticoid receptor-dependent trans-repression of activator protein-1. Mol Endocrinol. 2008;22(8):1767–80. doi: 10.1210/me.2007-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diefenbacher ME, et al. The LIM domain protein nTRIP6 recruits the mediator complex to AP-1-regulated promoters. PLoS One. 2014;9(5):e97549. doi: 10.1371/journal.pone.0097549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kassel O, et al. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-kappaB-regulated promoters. Genes Dev. 2004;18(20):2518–28. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diefenbacher ME, et al. The nuclear isoform of the LIM domain protein Trip6 integrates activating and repressing signals at the promoter-bound glucocorticoid receptor. Mol Cell Endocrinol. 2010;320(1–2):58–66. doi: 10.1016/j.mce.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 68.Yang L, et al. Interaction of the tau2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol Biol Cell. 2000;11(6):2007–18. doi: 10.1091/mbc.11.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chodankar R, et al. Hic-5 is a transcription coregulator that acts before and/or after glucocorticoid receptor genome occupancy in a gene-selective manner. Proc Natl Acad Sci U S A. 2014;111(11):4007–12. doi: 10.1073/pnas.1400522111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chodankar R, et al. Selective coregulator function and restriction of steroid receptor chromatin occupancy by Hic-5. Mol Endocrinol. 2015;29(5):716–29. doi: 10.1210/me.2014-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou H, et al. Frequency and distribution of AP-1 sites in the human genome. DNA Res. 2005;12(2):139–50. doi: 10.1093/dnares/12.2.139. [DOI] [PubMed] [Google Scholar]
- 72.Wiench M, et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30(15):3028–39. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanson RW, Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu Rev Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 74.Lin B, Morris DW, Chou JY. Hepatocyte nuclear factor 1alpha is an accessory factor required for activation of glucose-6-phosphatase gene transcription by glucocorticoids. DNA Cell Biol. 1998;17(11):967–74. doi: 10.1089/dna.1998.17.967. [DOI] [PubMed] [Google Scholar]
- 75.Schmoll D, Allan BB, Burchell A. Cloning and sequencing of the 5′ region of the human glucose-6-phosphatase gene: transcriptional regulation by cAMP, insulin and glucocorticoids in H4IIE hepatoma cells. FEBS Lett. 1996;383(1–2):63–6. doi: 10.1016/0014-5793(96)00224-4. [DOI] [PubMed] [Google Scholar]
- 76.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159(6):1327–40. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–26. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51(3):310–25. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang DX, Glass CK. Towards an understanding of cell-specific functions of signal-dependent transcription factors. J Mol Endocrinol. 2013;51(3):T37–50. doi: 10.1530/JME-13-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ou CY, et al. Coregulator cell cycle and apoptosis regulator 1 (CCAR1) positively regulates adipocyte differentiation through the glucocorticoid signaling pathway. J Biol Chem. 2014;289(24):17078–86. doi: 10.1074/jbc.M114.548081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu DY, et al. Distinct, genome-wide, gene-specific selectivity patterns of four glucocorticoid receptor coregulators. Nucl Recept Signal. 2014;12:e002. doi: 10.1621/nrs.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]