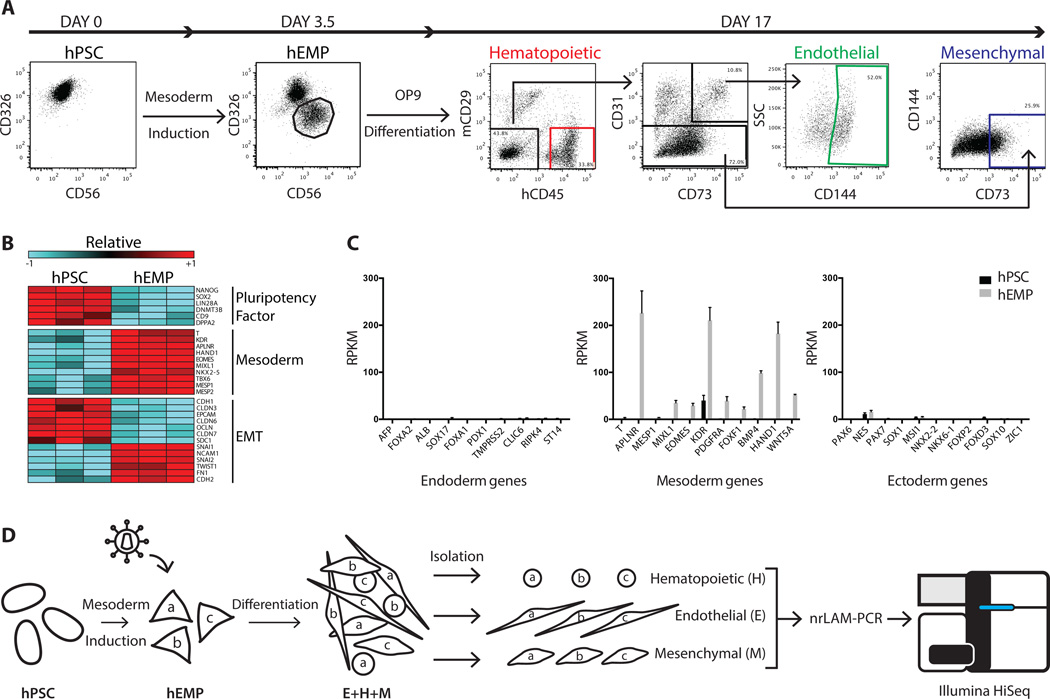
Figure 1.
Generation of a human embryonic mesodermal progenitor population from hPSC with hematopoietic, endothelial, and mesenchymal potential. (A): Flow cytometry analysis of EPCAM/CD326 and NCAM/CD56 expression in undifferentiated hPSC and hEMP generated after 3.5 days of sequential morphogen induction with Activin A, BMP4, VEGF and bFGF. hEMP were FACS isolated as the CD326−CD56+ population. Further differentiation on the murine stromal line OP9 generated hematopoietic, endothelial, and mesenchymal cells after 2 weeks. After exclusion of mCD29+ murine cells, the cell surface marker CD45 was used to define human hematopoietic cells. The endothelial lineage was isolated by the markers CD144+CD31+CD73+CD45−, a phenotype that defines non-hemogenic endothelium [14]. The mesenchymal lineage was isolated by the markers CD73+CD31−CD45−CD144−. (B): Transcriptome comparison via RNA-Seq of hPSC and hEMP showed downregulation (blue) of pluripotency factors, upregulation (red) of mesoderm genes and changes in gene expression indicative of EMT. Both FDR < 0.01 and fold change > 2-fold were applied as filters. Color scale in the heatmap shows the relative expression for each gene using its min/max moderate expression estimates as reference. (C): Gene expression in hPSC (black) and hEMP (gray) showed marked upregulation of mesodermal genes enrichment in hEMP without ectoderm or endoderm gene expression. (D): Schematic of lentiviral tagging approach to interrogate lineage potential of hEMP. After mesodermal induction of hPSC, hEMP were isolated at day 3.5, and transduced with a lentiviral vector. For the purposes of illustration, three distinct lentiviral tags created by transduction are shown as “a,” “b,” and “c.” These transduced hEMP were cocultured on OP9 stroma with conditions supporting differentiation of hematopoietic, endothelial, and mesenchymal cells; these lineages as well as lin neg cells were FACS isolated after two weeks (as in Fig. 1A). Integration sites in each lineage (E, H, and M) were identified by nrLAM-PCR followed by high throughput sequencing on an Illumina HiSeq. Abbreviations: EMT, epithelial-mesenchymal transition; hEMP, human embryonic mesoderm progenitor; hPSC, human pluripotent stem cell; nrLAM-PCR, non-restrictive linear amplification-mediated PCR; RPKM, Reads Per Kilobase of transcript per Million mapped reads.