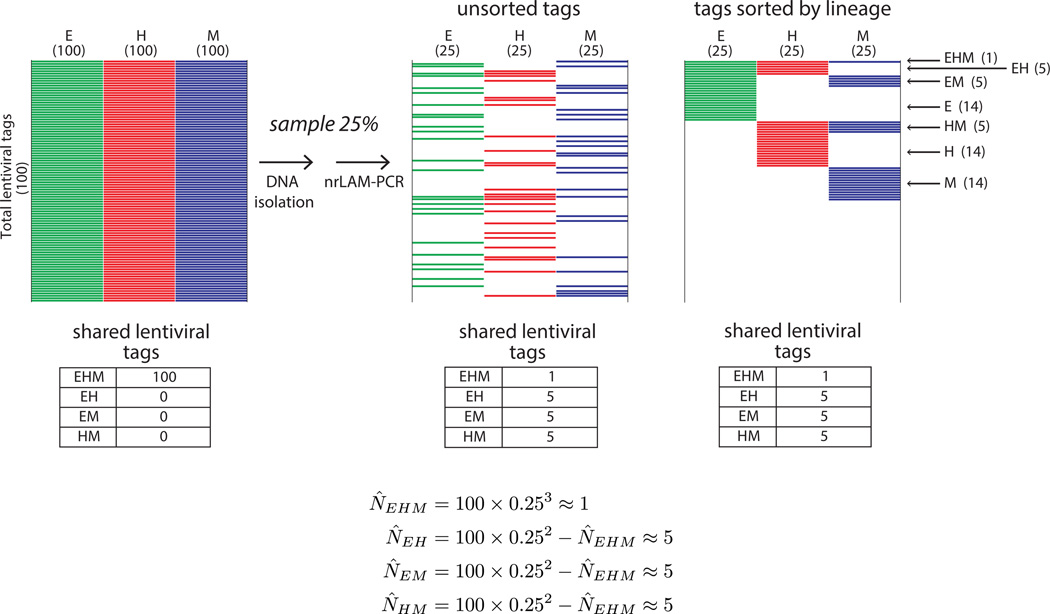
Figure 3.
Theoretical effect of sampling on shared lentiviral tag detection. Schematic heatmaps are shown for the theoretical scenario of a population in which all cells are multipotent and marked by a total of 100 lentiviral tags. Each multipotent cell goes on to form E, H, and M progeny, and all three lineages therefore contain all 100 tags in the culture vessel (left panel). We show how the results of this experiment appear using a model in which only 25% of tags are found from each lineage because of cumulative undersampling during cell and DNA processing. The tags are unsorted in the middle panel to reflect the randomness of the sampling; in the right panel the same tags are sorted according to lineage to aid visualization. With this degree of undersampling (25%), an average experiment will only detect 58 of the 100 lentiviral tags, only 15 will be shared by two lineages, and most importantly, only one tag will be detected as shared between the three lineages. Abbreviations: E, endothelial; H, hematopoietic; M, mesenchymal; nrLAM-PCR, non-restrictive linear amplification-mediated PCR.