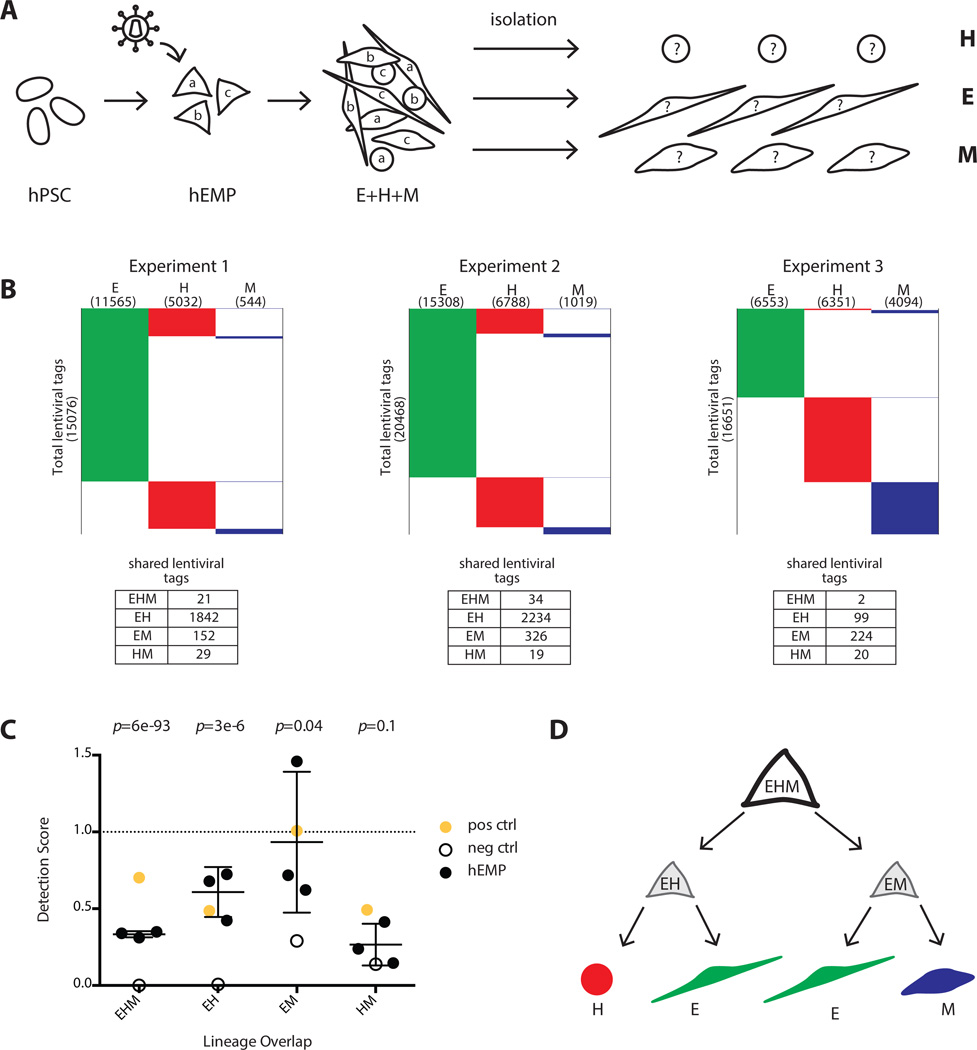
Figure 5.
Lentiviral tagging of human embryonic mesodermal progenitors reveal subpopulations with bipotent and tripotent potential. (A): Experimental design to track clonal output of a pool of tagged mesoderm progenitors (hEMP) differentiated into H, E, and M lineages. (B): Data from three independent experiments showing number of lentiviral tags identified in each lineage. Experiment 1 was performed with the integrase inhibitor raltegravir added after 12 hour of transduction while experiments 2 and 3 were performed without raltegravir. The addition of raltegravir did not alter the detection score of shared lentiviral tags. (C): The detection scores from marking mesoderm progenitors are shown (black dots) as well as the maximum detection expectation (set by positive control, yellow dot) and the false positive rate (set by negative control, empty circle). The negative control detection scores for EHM, EH, and EM tag sharing were significantly lower than the detection scores in the mesoderm progenitor tagging experiments (p-values 6 × 10−93, 3 × 10−6 and .04, respectively for EHM, EH, and EM). The HM detection scores were similar between the progenitor tagging experiments and the negative control (p-value = .1). p-value calculation used an extreme value test performed using the cumulative distribution function of a normal distribution with mean and standard deviation calculated from progenitor experimental data. (D): Proposed model of the developmental potential of human mesodermal progenitor defined by unbiased lentiviral marking. During mesodermal differentiation from hPSC, a tripotent progenitor (EHM) with limited self-renewal ability rapidly generates either endothelial/hematopoietic progenitors (EH) or endothelial/mesenchyme progenitor (EM). The lentiviral tagging data do not support the existence of a bipotent HM progenitor. Abbreviations: E, endothelial; H, hematopoietic; hEMP, human embryonic mesoderm progenitor; hPSC, human pluripotent stem cell; M, mesenchymal.