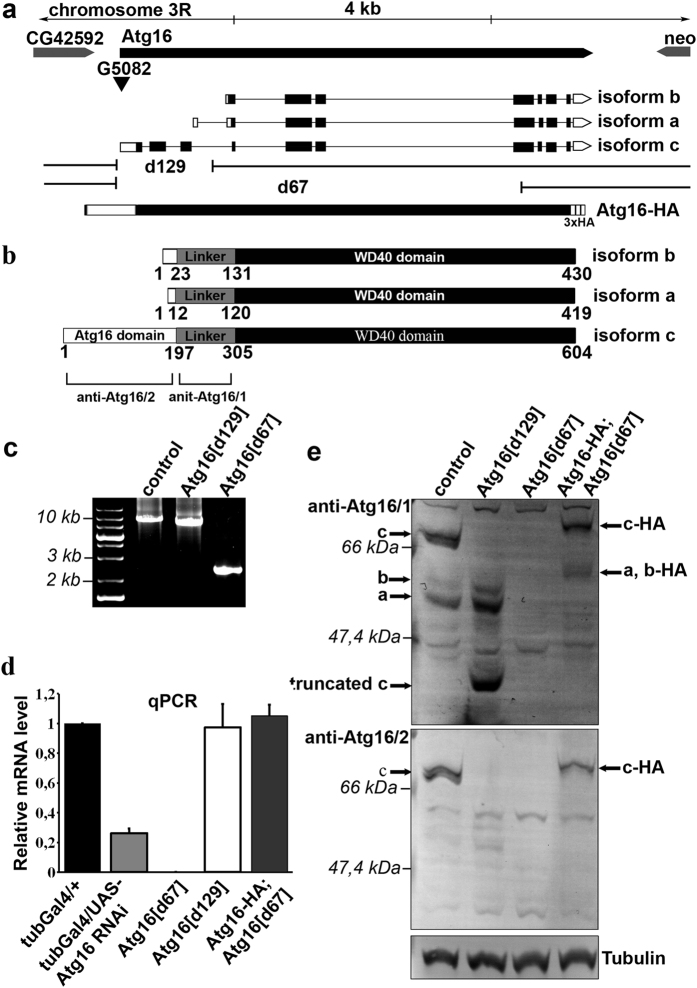
Figure 1. Generation of Atg16 mutations, rescue transgene, and antibodies.
(a) Two Atg16 mutations were isolated by improper excision of the P element G5082, which is located in the 5′ UTR of this gene. Atg16d129 carries a 1,977 bp deletion removing the first part of the Atg16 gene, and Atg16d67 contains a 7,914 bp deletion removing much of the gene. The composition of the genomic promoter-driven Atg16-HA transgene is also shown. The Atg16 mRNA isoforms are indicated, with open bars representing untranslated regions and closed bars representing coding sequences. (b) This panel illustrates the domain structures of Atg16 protein isoforms, and the recognition regions of anti-Atg16/1 and 2. (c) PCR amplification from genomic DNA with primers specific for flanking regions of the Atg16 gene shows the size of the deletions in Atg16d129 and Atg16d67 adult flies compared to control. (d) Quantitative real-time PCR shows a decrease of Atg16 mRNA level in Atg16 RNAi and Atg16d67 mutant flies compared to control and rescued Atg16d67 mutants. No decrease of mRNA expression is seen in Atg16d129 mutants because primers are specific for a common region of the 3 mRNA isoforms. Standard error values are ±0.031131; 0.000001; 0.0156954; 0.052920 (the tub-Gal4/ + control was set to 1). (e) Western blots using linker region-specific (anti-Atg16/1) and Atg16 domain-specific (anti-Atg16/2) antibodies show the expression of the various Atg16 isoforms in control, Atg16d129 and Atg16d67 mutants, and in Atg16d67 mutants rescued by Atg16-HA.