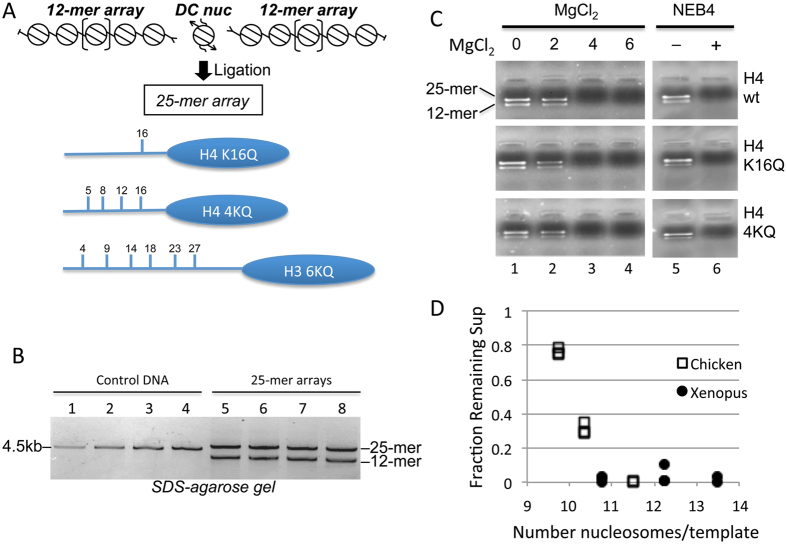
Figure 1. Histones and nucleosome arrays.
(A) Positions of lysine acetylation mimics and ligation strategy. Top, Schematic showing arrangement of 12-mer arrays and DC nucleosome upon ligation to the 25-mer nucleosome array. Directional ligation sites are indicted by arrowheads, t-bars indicate unligatable DNA ends. Bottom. The locations of lysine to glutamine substitutions within H3 and H4 used in this study are shown. (B) Increasing amounts of the 4.5 kb control DNA (1, 2, 3, and 4 μg) are loaded in lanes 1–4. Ligated WT, H4 4KQ, H3 6KQ and H3 6KQ/H4 4KQ arrays are shown (lanes 5–8). Some unligated 12-mer array remains after ligation. (C) Ligated arrays are saturated with nucleosomes and completely self-associated in NEB4 digestion buffer. Self-association assays with the arrays (right) were performed as described with increasing MgCl2+ (lanes 1–4), as indicated. Self-association of arrays in the absence (−) or presence (+) of NEB4 buffer. (D) Self-association and analytical ultracentrifugation (AU) analysis of arrays in NEB4 buffer. 12-mer nucleosome array templates were reconstituted to increasing saturations with either chicken or recombinant Xenopus core histones and the level of saturation determined by AU. The fraction of each array remaining in solution as determined by self-association assays was plotted vs number of nucleosomes per template. Duplicates of assays are shown, 12 nucleosomes per template is considered saturation.