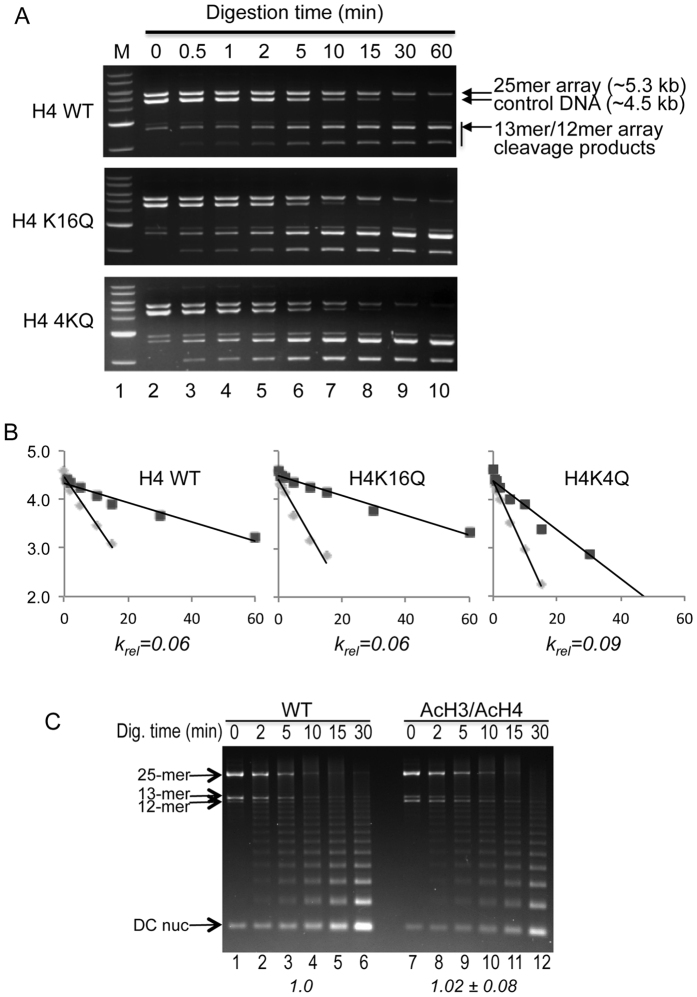
Figure 2. Accessibility of the central nucleosome linker DNA within a 25-nucleosome array.
(A) Example of digestion time course for arrays containing wt H4, H4 K16Q or H4 4KQ within the central DC nucleosome. Time points (min.) of digestion are shown at the top of the gel. Digestion products were analyzed on 0.7% SDS-agarose gels followed by EtBr staining. Bands corresponding to the 5.3 kb 25-mer template, the 4.5 kb naked DNA control, and residual 12/13 mer arrays and other digestion products are indicated. Lane 1 contains molecular size markers (M). (B) Example plots of quantified digestion data as described in Materials and Methods. Lines represent linear regression fits with R2 for each fit ranging from 0.96 to 0.98. The rate of the chromatin digestion relative to the naked DNA internal control (krel) is indicated below each plot. (C) Effects of acetylation mimics are localized to the DC nucleosome in the 25-mer arrays. The 25-mer arrays were digested with Eco RI and products run on an SDS-agarose gel. Note that EcoRI sites lie between each nucleosome in the 25-mer array. Relative rate of the loss of the uncut 25-mer normalized to WT is indicated below the gel.