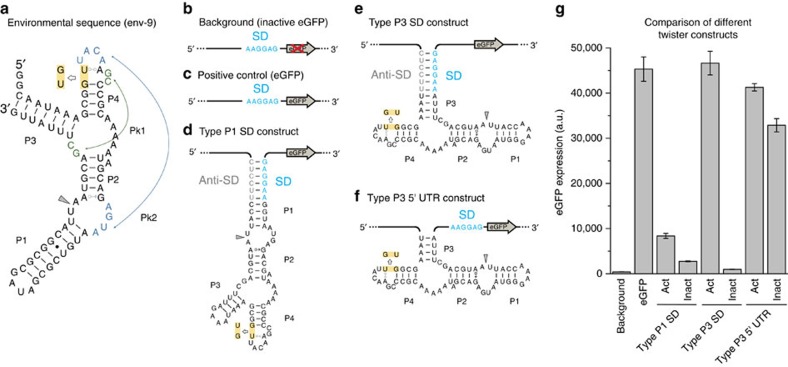
Figure 1. The env-9 twister ribozyme motif and its engineered 5′-UTR constructs in Escherichia coli.
The env-9 motif can be used to control gene expression at the translational level in E. coli. (a) Sequence and secondary structure of the natural occurring env-9 motif. The pseudoknots Pk1 and Pk2 are highlighted in green and blue, respectively. The inactivating mutation is highlighted in yellow. The cleavage site is indicated with a grey arrow. A negative control carrying a truncated form of eGFP (b) was used to measure the background fluorescence of the culture. A plasmid carrying the active form of eGFP (c) was used as a positive control. The Shine–Dalgarno sequence (SD) is shown in light blue. (d–f) The three engineered constructs inserted in the 5′-UTR of the eGFP gene. In the type P1 SD construct (d) the SD (in blue) and the anti-SD (in grey) sequences were included into stem P1 of the twister. In the type P3 SD construct (e) the SD and the anti-SD sequence were placed into stem P3. In the type P3 5′-UTR construct (f) the twister motif is inserted in the 5′-UTR before the SD of the eGFP gene (see Supplementary Fig. 1 for details). In the constructs shown in e,f the stem P1 was shortened and closed with a 5′-GAAA-3′ tetraloop. (g) The levels of eGFP expression (bulk fluorescence divided by the relative OD600) of the constructs displayed in b–f are shown. The error bars represent s.d. calculated on independent biological triplicates.