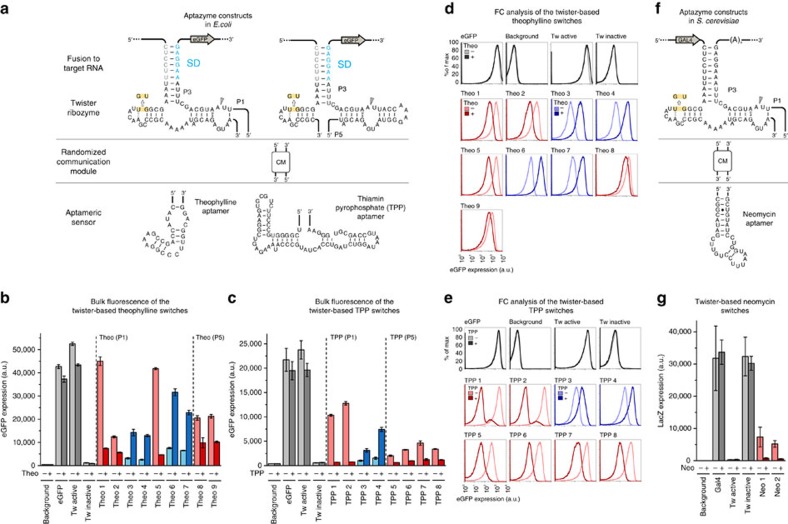
Figure 2. Twister-based one-input riboswitches in Escherichia coli and Saccharomyces cerevisiae.
(a) Theophylline and TPP aptazyme design in E. coli. Different types of communication modules (CM) were employed (see Supplementary Figs 4 and 5). The Shine-Dalgarno (SD) and the anti-SD are shown in light blue and grey, respectively. The inactivating mutation is highlighted in yellow. The cleavage site is indicated with a grey arrow. (b,c) Level of eGFP expression (bulk fluorescence divided by the relative OD600) of the selected clones in the absence (−) and in the presence (+) of (b) 2.5 mM theophylline and (c) 1 mM thiamin in the culture medium. The levels of reporter gene expression in the different conditions are represented by grey tone bars for the controls, red tone bars for the off-switches and blue tone bars for the on-switches. The error bars represent s.d. calculated on independent biological triplicates. (d,e) flow cytometry (FC) histograms of (d) the theophylline switches and (e) the TPP switches recorded in the presence (+) and in the absence (−) of the respective ligands (2.5 mM theophylline and 1 mM thiamin in the culture medium respectively). The FC diagrams of the controls are represented by grey tone traces, the diagrams of the off-switches by red tone traces and the ones of the on-switches by blue tone traces. (f) Neomycin aptazyme design in S. cerevisiae. Here the aptazyme was inserted into the 3′-UTR of the GAL4 gene. The inactivating mutation is highlighted in yellow, the cleavage site is indicated with a grey arrow. (g) Levels of the reporter gene expression (LacZ), defined as chemoluminescence divided by the relative OD600, of the selected clones in the absence (−) and in the presence (+) of 100 μg ml−1 neomycin in the culture medium. The levels of reporter gene expression in the different conditions are represented by grey tone bars for the controls and red tone bars for the off-switches. The error bars represent s.d. calculated on independent biological triplicates.