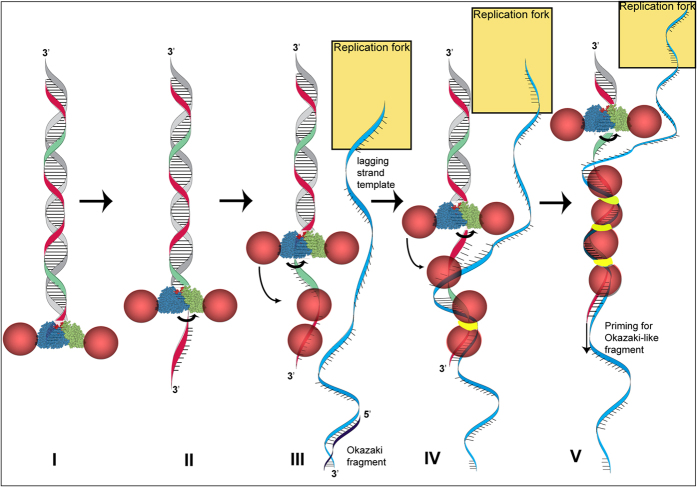
Figure 6. Model for Redαβ molecular crosstalk.
(I) Redα toroids (blue and green cones) thread onto the 3′ end of a dsDNA break (I) and digest nucleotides from the 5′ end, extruding the 3′ end as single stranded (red strand; (II)). Redβ monomers (pink spheres) bound to Redα are promoted to bind to the emerging ssDNA (III). Upon locating complementary sequence on the lagging strand template (blue strand) at the replication fork, a Redβ dimer undergoes a conformational change (yellow band) to establish a DNA clamp (IV) that nucleates nucleoprotein filament growth and allows, possibly promotes, the 3′ end (red strand) to serve as a primer for Okazaki-like fragment synthesis (V). The replication fork is depicted as a golden rectangle.