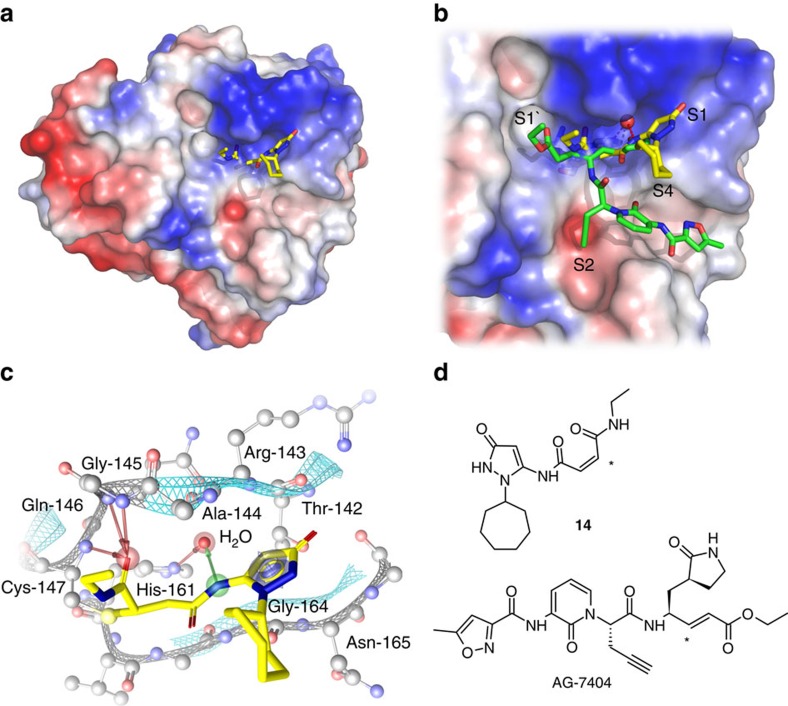
Figure 6. Crystal structure of inhibitor 14 with 3C protease of EV-B93 (PDB: 5IYT).
(a) Refined structure of the enzyme–inhibitor complex with 14 represented as sticks, carbon atoms in yellow. (b) Close-up of the substrate-binding site with bound 14 and AG-7404 (carbon atoms in green); sub-pockets S1', S1, S2 and S4 are annotated. Water molecules are represented as red spheres. (c) Binding modes of 14; EV-B93 3C amino-acid residues in close proximity to 14 are labelled in the S1 and S1' sub-pockets. The arrows represent the H-bonds between 14, water molecules crucial for inhibitor binding and the protein. (d) Structures of inhibitors 14 and AG-7404 with the carbon covalently attacked by the sulfur nucleophile of the protease being highlighted by an asterisk.