Abstract
Metagenomic analysis of the human intestinal microbiome has provided a wealth of information that allowed an exceptionally detailed description of its microbial content and physiological potential. It also set the basis for studies allowing correlation of alterations in the balance of this microbiota and the occurrence of a certain number of emerging diseases, such as inflammatory bowel diseases, obesity and diabetes, and possibly colorectal cancer. The time has come to give the intestinal microbiota in symbiosis with its host an experimental dimension. This brief review summarizes our attempt at developing a cellular microbiology of the mutualistic symbiosis established between the gut microbiota and the host intestinal surface. Particular attention is paid to the intestinal crypt, due to its role in epithelial regeneration.
This article is part of the themed issue ‘The new bacteriology’.
Keywords: gut microbiota, intestinal crypts, pathobiont
1. The gut microbiota in symbiosis with humans
The human gut, particularly the colon, is host for about a thousand bacterial species that together represent the astronomic number of about 1014 individual microorganisms called the gut microbiota. The major rules directing the constitution of this microbiota following birth, its balance and maintenance, as well as its resilience in case of a ‘catastrophic’ event such as administration of an antibiotic, have been increasingly addressed over the past decades [1,2]. The dominance of Firmicutes and Bacteroidetes was established although it was soon realized that a significant proportion of the species composing the gut microbiota could not be cultivated. It took the first 16S DNA analysis [3] to realize that the microbial diversity was even higher than anticipated. In the same period, it also became obvious that the gut microbiota is in a mutualistic interaction with its host. This is particularly the case regarding the maturation and controlled activation (i.e. physiological inflammation) of the mucosal and systemic immune system [4–7], nutrition and metabolism, particularly the capacity offered by the gut microbiota to digest and ferment complex plant polysaccharides [8–10].
However, following the sequencing of the human genome [11], it became obvious that attention needed to be paid to the human hologenome, in other words its eukaryotic and prokaryotic genetic patrimony [12]. Thus, in 2010, the almost 10-year due metagenome of the gut microbiota of more than a hundred individuals was eventually published following completion of the EU-funded MetaHIT programme [13]. Using massive shot-gun metasequencing, the consortium identified a set of about 3.3 million prokaryotic genes constituting the global prokaryotic genome patrimony among the set of studied individuals which has now been expanded to a pan-metagenome of about 10 million genes among which we, as humans, are likely to share a small portion (about 536 000 genes) representing the core metagenome involved in the basic mutualistic interaction. Metagenomic analysis, in its global dimension, enables the topic of microbiota to be taken away from the concept of an assemblage of microbes which, in their extraordinary interhuman diversity and variation, defy our capacity to address the actual functional logic of the symbiosis, and to address the latter question in terms of genes/operons functions, thus providing a more physiological view of the interface [14–16].
Still, metagenomics remains essentially descriptive, enabling the identification of a set of core functions, or to possibly reassemble the constituent genomes and the nature and quantity of an increasing number of species. Metagenomics can also be correlative, as an increasing number of researchers establish strong associations between loss of balance of the microbiota (i.e. dysbiosis) and certain pathological conditions, including obesity and diabetes, inflammatory bowel diseases (IBDs) and colon cancer [17]. Metagenomics, to some extent, can also be made experimental, for instance, when studying its resilience following antibiotic treatment, or its alterations following diet modifications [18,19].
2. Commensal bacteria in the murine gut
Our aim is to bring symbiosis to a level of molecular and cellular analysis, in other words, to capitalize on the expertise acquired in deciphering pathogenic mechanisms, also called cellular microbiology, to decipher the molecular cross-talk that characterizes the mutualistic signals exchanged between symbiotic microbes and intestinal cells and tissues. It is a complex endeavour because the genetics of the relevant microorganisms are rarely established and the phenotypes of interaction (i.e. symbiosis endpoints and readouts) are often not as clearly characterized as those of pathogenic interactions (i.e. cell adhesion/invasion, cell death, inflammation/destruction of tissues, etc.) [20,21].
We have entered the period of ‘experimentomics’ and accordingly set a series of research lines that address the diversity in developing simplified models of interaction.
In brief, following the development of a method based on whole-genome ‘signature-tagged mutagenesis’ [22], in Lactobacillus casei, we identified 47 genes that appeared essential for establishment of this symbiont in the gut [23]. The library encompassing 1100 individual and identifiable mutants can also be used for other mechanistic studies such as in a model system allowing the study of the impact of symbionts on nutrient transepithelial transport, particularly lipids.
We have also established the proper conditions to achieve in vitro culture of Candidatus arthromitus, the segmented filamentous bacterium (SFB), belonging to the Clostridiale family that had resisted culture attempts for 50 years [24]. This ‘pathobiont’ seems essential following weaning in mammals to programme the infant's immune system, particularly the differentiation of naive T cells into Th1, Th17 and Th2 lymphocytes [25]. It was thus essential to establish the proper conditions of microbe–host interaction in order to address the molecular cross-talk leading to immune maturation. Reverse genetics of SFB is also on its way.
3. Commensal bacteria in murine intestinal crypts
Last but not least, we have selected the intestinal crypt as a model to study microbiota–host cross-talk. Our starting hypothesis was that among the complex assemblage of luminal and mucosal commensal species that compose the microbiota, a limited set may enter into a mutualistic interaction that may reflect a long coevolution that established a situation in which the epithelial regenerative apparatus may benefit from microbiota-mediated protection.
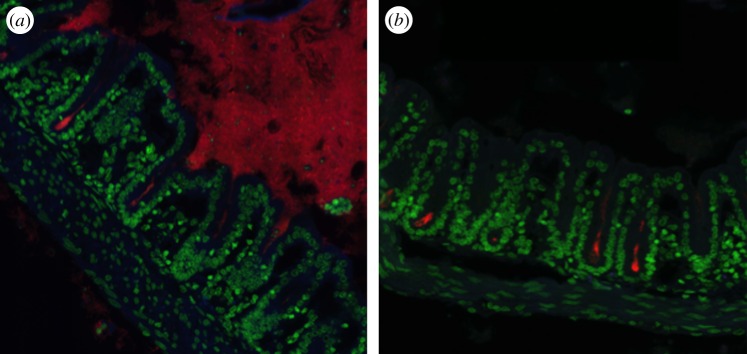
At stake is the homeostasis of a particularly sensitive zone where adult stem cells are directly exposed to the gut flora. Preliminary evidence collected from Warthin–Starry (silver nitrate) staining of various segments of the intestinal tract of various mouse lines reproducibly showed the presence of a small cluster of bacteria located at the crypt bottom in the caecum and proximal colon. These bacteria were not seen in general in the duodeno-jejunum and in the distal colon. They were positively marked by FISH using a universal 16S rRNA probe, indicating that these bacteria were alive and metabolically active (figure 1). We next developed a dedicated pipeline to molecularly identify the relevant bacteria. Using a combination of laser capture microdissection of the crypt luminal space on intestinal tissue section, amplification of the variable sequences of genomic DNA encoding 16S rRNA, next-generation sequencing (454 pyrosequencing) and bioinformatic analysis, we could demonstrate that these bacterial clusters were dominated by operational taxonomic units (OTUs) corresponding to totally unexpected commensal microorganisms such as Acinetobacter and Delftia/Comamonas, in other words strictly aerobic, non-fermentative genera, respectively, belonging to the gamma- and beta-proteobacterial families [26]. We could subsequently cultivate several isolates constituting the so-called ‘crypt-specific core microbiota’ (CSCM) and further identify them as mostly A. parvus, A. radioresistens and D. acidovorans. Following gavage of axenic mice with the relevant Acinetobacter species, we have recently confirmed their consistent tropism to the crypts, thus questioning the nature of the parameters dictating the presence and restriction of Acinetobacter to this particular niche. It was similarly described that following mono-association of germ-free mice with the bona fide commensal species Bacteroides fragilis, colonic crypts became colonized, thereby identifying a sugar transport system as a key element for bacterial survival in this hard-to-explore environment [27]. The presence of a low tension of O2 at the surface of the intestinal epithelium was demonstrated [28] and could partly account for the selection of these strictly aerobic microorganisms. Other parameters are likely to be involved. It also becomes essential to understand the rationale for the presence of a CSCM in this very sensitive zone that is critical for epithelial regeneration. Our central hypothesis is that the CSCM may act as a crypt ‘gate-keeper’ with multiple complementary functions selected by the coevolutionary process: (i) protection against the intrusion of pathogens or pathobionts that may disturb the fragile homeostasis required to preserve the balance of epithelial regeneration in physiological conditions or following a cytotoxic aggression, and ‘buffering’ of inflammation that may be transiently caused by the accidental passage of a pro-inflammatory pathobiont. (ii) Biodegradation of the xenobiotic molecules that may gain access to the crypt and induce strong genotoxic damage, particularly on stem cells. It must be noted that the CSCM isolates are typical environmental microorganisms with strong and diverse biodegradative activities, as assessed by the annotation of their genomic sequences [29].
Figure 1.
Fluorescence in situ hybridization of murine proximal colonic tissue with an Alexa-555 pan bacteria probe (a) or with an Alexa-555 Acinetobacter specific probe (b). Nuclei in green are stained with DAPI. Pictures were taken with an Opterra system (Bruker) using a 63× objective.
Our current work is aimed at deciphering the logic of the CSCM-crypt symbiosis and to extend this concept to other mammalian species, including humans. At stake is the better understanding of the pathogenic mechanisms of IBDs and colon cancer.
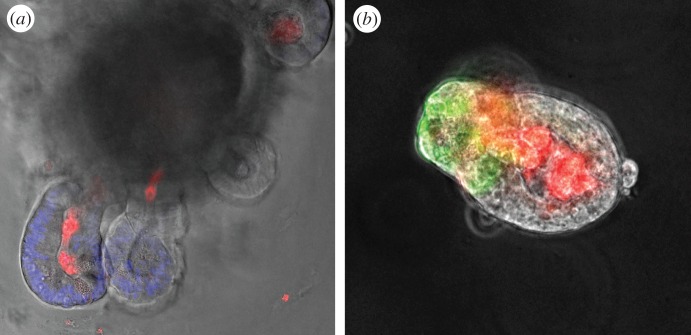
In this context, it was clear that our hypothesis also needed to be validated by demonstrating the existence of a true cross-talk between the microbiota, possibly more specifically the CSCM and the crypt. For this, we developed an ex-vivo model of interaction based upon the recent work of Hans Clever's group: the ‘miniguts’ or ‘organoids’ [30]. Following purification of murine intestinal crypts and their embedding in Matrigel in the presence of essential growth factors such as R-Spondin, Noggin and Wnt ligands, organoids can be grown and maintained in vitro, where they display the progressive appearance of crypt-like structures composed of stem cells, as well as a transit-amplifying compartment followed by cell cycle arrest and differentiation into the various epithelial lineages: Paneth cells in close apposition to stem cells, bona fide epithelial cells, goblet cells and enterochromaffin cells [31,32]. When crypts were exposed, before embedding, to bacterial MAMPs (microbe-associated molecular patterns), such as peptidoglycan (PGN) muramyl-dipeptide (MDP), muramyl-tri and tetrapeptide, lipopolysaccharide, flagellin and lipoproteins (Pam3CSK), only crypts exposed to PGN and MDP yielded four- to fivefold more organoids compared with unstimulated organoids (figure 2). Further experiments indicated that Lgr5+/CD24middle+ crypt cells corresponding to the stem cells expressed high levels of NOD2 transcripts, the intracellular cytosolic sensor involved in MDP recognition. This enabled the identification of an MDP–NOD2 pathway of stem cell cytoprotection in the murine intestinal epithelium [33].
Figure 2.
Typical images of intestinal organoids treated with MDP-rhodamine, showing that the MDP is internalized in the lumen of the organoid during the embedding process. In (a) organoid from wild-type mouse (nuclei are stained in blue with DAPI and MDP in red) and in (b) from lgr5-EGFP mouse (stem cells are in green and MDP in red).
Moreover, when the organoids were generated from crypts extracted from mice pretreated for 72 h with doxorubicin, a potent cytotoxic DNA-intercalating drug causing massive oxidative stress response that appears particularly cytotoxic to stem cells, the yield of organoids following MDP stimulation was increased by a factor of 15 and more, thereby indicating that the MDP–NOD2 pathway is more protective following cytotoxic aggression than in homeostatic conditions. In addition, the index of epithelial regeneration following cytotoxicity was strongly increased in wild-type conventional mice compared with NOD2−/− mice.
Altogether this provides strong support to the concept of a protective cross-talk between the microbiota and the regenerative apparatus of the crypt with particular targeting to the stem cells.
4. Conclusion
Our approach has attempted to set the basis for a cellular microbiology of the mutualistic symbiosis established between elements of the intestinal microbiota and the gut mucosal tissues. Three examples were selected which do not reflect the wealth of different situations encompassing this rich symbiosis. Particular attention has been paid to defining the basic parameters of gut colonization by a symbiont; to cultivating a key pathobiont that stimulates maturation of the innate/adaptive immune system following weaning; and to unravelling the cross-talk established in the intestinal crypt between bacterial MAMPs, such as MDP, and stem cells. Moreover, in order to decipher the factors allowing Acinetobacter to express its tropism for colonic crypts, we will apply a reverse genetics approach to identify genes that are essential for colonization of this particular niche. A natural extension of our work will be to study if a CSCM is also present in human colonic crypts, both in healthy and pathological states.
Authors' contributions
T.P., G.N. and P.J.S. wrote the paper.
Competing interests
We have no competing interests.
Funding
This work was supported by the European Research Council (P.J.S. Advanced Grants 339579-DECRYPT). P.J.S. is a Howard Hughes Medical Institute Foreign Scholar.
References
- 1.Savage DC. 2001. Microbial biota of the human intestine: a tribute to some pioneering scientists. Curr. Issues Intest. Microbiol. 2, 1–15. [PubMed] [Google Scholar]
- 2.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. ( 10.1038/nature11550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manichanh C, et al. 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211. ( 10.1136/gut.2005.073817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberl G, Lochner M. 2009. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2, 478–485. ( 10.1038/mi.2009.114) [DOI] [PubMed] [Google Scholar]
- 5.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. ( 10.1038/nri2515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336, 1268–1273. ( 10.1126/science.1223490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157, 121–141. ( 10.1016/j.cell.2014.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl Med. 1, 6ra14. ( 10.1126/scitranslmed.3000322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut. Microbes 3, 289–306. ( 10.4161/gmic.19897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504. ( 10.1038/nrmicro3050) [DOI] [PubMed] [Google Scholar]
- 11.McPherson JD, et al. 2001. A physical map of the human genome. Nature 409, 934–941. ( 10.1038/35057157) [DOI] [PubMed] [Google Scholar]
- 12.Rosenberg E, Sharon G, Zilber-Rosenberg I. 2009. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ. Microbiol. 11, 2959–2962. ( 10.1111/j.1462-2920.2009.01995.x) [DOI] [PubMed] [Google Scholar]
- 13.Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. ( 10.1038/nature08821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon MY, Lee KM, Yoon Y, Go J, Park Y, Cho YJ, Tannock GW, Yoon SS. 2013. Functional screening of a metagenomic library reveals operons responsible for enhanced intestinal colonization by gut commensal microbes. Appl. Environ. Microbiol. 79, 3829–3838. ( 10.1128/AEM.00581-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang WL, Xu SY, Ren ZG, Tao L, Jiang JW, Zheng SS. 2015. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 21, 803–814. ( 10.3748/wjg.v21.i3.803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arumugam M, et al. 2011. Enterotypes of the human gut microbiome. Nature 473, 174–180. ( 10.1038/nature09944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Althani A, Marei H, Hamdi WS, Nasrallah GK, El Zowalaty ME, Al Khdor S, Al-Asmakh M, Abdel-Aziz H. 2015. Human microbiome and its association with health and diseases. J. Cell Physiol. 231, 1688–1694. ( 10.1002/jcp.25284) [DOI] [PubMed] [Google Scholar]
- 18.Fitzpatrick D, Walsh F. 2016. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol. Ecol. 92, fiv168. ( 10.1093/femsec/fiv168) [DOI] [PubMed] [Google Scholar]
- 19.Quercia S, et al. 2014. From lifetime to evolution: timescales of human gut microbiota adaptation. Front. Microbiol. 5, 587 ( 10.3389/fmicb.2014.00587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sansonetti PJ. 2004. War and peace at mucosal surfaces. Nat. Rev. Immunol. 4, 953–964. ( 10.1038/nri1499) [DOI] [PubMed] [Google Scholar]
- 21.Sansonetti PJ. 2011. To be or not to be a pathogen: that is the mucosally relevant question. Mucosal Immunol. 4, 8–14. ( 10.1038/mi.2010.77) [DOI] [PubMed] [Google Scholar]
- 22.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403. ( 10.1126/science.7618105) [DOI] [PubMed] [Google Scholar]
- 23.Licandro-Seraut H, Scornec H, Pédron T, Cavin JF, Sansonetti PJ. 2014. Functional genomics of Lactobacillus casei establishment in the gut. Proc. Natl Acad. Sci. USA 111, E3101–E3109. ( 10.1073/pnas.1411883111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. 2015. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103. ( 10.1038/nature14027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. 2013. Host interactions with segmented filamentous bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin. Immunol. 25, 342–351. ( 10.1016/j.smim.2013.09.001) [DOI] [PubMed] [Google Scholar]
- 26.Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. 2012. A crypt-specific core microbiota resides in the mouse colon. MBio 3, e00116-12. ( 10.1128/mBio.00116-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2011. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429. ( 10.1038/nature12447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marteyn B, et al. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465, 355–358. ( 10.1038/nature08970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saffarian A, Mulet C, Naito T, Bouchier C, Tichit M, Ma L, Grompone G, Sansonetti PJ, Pédron T. 2015. Draft genome sequences of Acinetobacter parvus CM11, Acinetobacter radioresistens CM38, and Stenotrophomonas maltophilia BR12, isolated from murine proximal colonic tissue. Genome Announc. 3, e01089-15 ( 10.1128/genomeA.01089-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. ( 10.1038/nature07935) [DOI] [PubMed] [Google Scholar]
- 31.Sato T, Clevers H. 2013. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194. ( 10.1126/science.1234852) [DOI] [PubMed] [Google Scholar]
- 32.Clevers H, Loh KM, Nusse R. 2014. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 ( 10.1126/science.1248012) [DOI] [PubMed] [Google Scholar]
- 33.Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ. 2014. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe. 15, 792–798. ( 10.1016/j.chom.2014.05.003) [DOI] [PubMed] [Google Scholar]