Abstract
Tuberculosis remains a scourge of global health with shrinking treatment options due to the spread of drug-resistant strains of Mycobacterium tuberculosis. Intensive efforts have been made in the past 15 years to find leads for drug development so that better, more potent drugs inhibiting new targets could be produced and thus shorten treatment duration. Initial attempts focused on repurposing drugs that had been developed for other therapeutic areas but these agents did not meet their goals in clinical trials. Attempts to find new lead compounds employing target-based screens were unsuccessful as the leads were inactive against M. tuberculosis. Greater success was achieved using phenotypic screening against live tubercle bacilli and this gave rise to the drugs bedaquiline, pretomanid and delamanid, currently in phase III trials. Subsequent phenotypic screens also uncovered new leads and targets but several of these targets proved to be promiscuous and inhibited by a variety of seemingly unrelated pharmacophores. This setback sparked an interest in alternative screening approaches that mimic the disease state more accurately. Foremost among these were cell-based screens, often involving macrophages, as these should reflect the bacterium's niche in the host more faithfully. A major advantage of this approach is its ability to uncover functions that are central to infection but not necessarily required for growth in vitro. For instance, inhibition of virulence functions mediated by the ESX-1 secretion system severely attenuates intracellular M. tuberculosis, preventing intercellular spread and ultimately limiting tissue damage. Cell-based screens have highlighted the druggability of energy production via the electron transport chain and cholesterol metabolism. Here, I review the scientific progress and the pipeline, but warn against over-optimism due to the lack of industrial commitment for tuberculosis drug development and other socio-economic factors.
This article is part of the themed issue ‘The new bacteriology’.
Keywords: antibiotics, drug resistance, drug discovery, phagocytes, tuberculosis
1. Introduction
The discovery of antibacterial agents and their application in human medicine have radically changed the course of infectious diseases, protected health and extended life expectancy [1]. Most life-threatening diseases can still be cured by antibiotic treatment but the range of therapeutic options offered by antibiotics is diminishing due to the emergence and spread of drug-resistant bacteria [2]. Drug resistance has arisen because of the overuse, misuse and abuse of antibiotics, and the transfer of drug resistance determinants between many bacteria. Alarmingly, the number of new antibiotics introduced into the clinic has shrunk to an all-time low as the pharmaceutical base has severely contracted and investment eroded [3].
Tuberculosis respects no boundaries and is a major, enduring threat to global health. The World Health Organisation reported 9.6 million incident cases in 2014, 12% of which were co-infected with HIV, and there were 1.5 million deaths of which 0.4 million were HIV-positive [4]. The burden of latent infection is estimated to be over 2 billion. Although the HIV epidemic appears to be receding there are grounds for concern with another potential co-morbidity, type 2 diabetes. This is predicted to affect 600 million individuals worldwide by 2050 and to be especially prevalent in developing countries such as India where there are currently 2.5 million active tuberculosis cases and approximately 60 million diabetics.
Tuberculosis, like other bacterial diseases, is confronted by problems of drug resistance. Treatment of drug-susceptible disease still comprises an intensive phase of two months with isoniazid, rifampicin, ethambutol and pyrazinamide followed by a four-month continuation phase of isoniazid and rifampicin alone, a regimen first devised 40 years ago. Multidrug-resistant tuberculosis (MDR-TB) arises when strains display resistance to isoniazid and rifampicin, whereas extensively drug-resistant tuberculosis (XDR-TB) also involves resistance to the second-line drugs, notably a fluoroquinolone and one of the injectables (capreomycin, kanamycin or streptomycin). In 2014, MDR-TB affected 3.3% of new cases and 20% of previously treated cases, and accounted for 190 000 deaths [4]. Overall, there were 480 000 cases of MDR-TB and an estimated 48 000 cases of XDR-TB. In the light of these staggering statistics there is clearly a need for new and more effective drugs.
2. Discovering new drugs and regimens
Since publication of the genome sequence of the H37Rv strain of Mycobacterium tuberculosis in 1998 [5], a variety of different genome-enabled approaches have been implemented to find new leads for drugs and to incorporate new drug candidates into regimens as multidrug therapy will remain mandatory for tuberculosis. These discovery efforts ranged from testing existing drugs for activity against M. tuberculosis to developing new ones ab initio. An important conceptual advance was the understanding of the necessity to begin testing new candidates in combinations during early stages of preclinical development in order to accelerate the subsequent clinical trials [6,7].
(a). Repurposing old drugs
In the 2000s, one means of accelerating the development of new treatment-shortening regimens was repurposing drugs that had proved successful in curing other infections. Particular interest was devoted to the newer fluoroquinolones as one of these, moxifloxacin, had proved highly effective in rapidly reducing the bacterial load in murine models of tuberculosis [8]. This, and observations from phase II trails [9], led to phase III clinical trials of moxifloxacin against pulmonary tuberculosis where the fluoroquinolone replaced one or other of the standard drugs in the frontline regimen with the expectation of faster cure. Disappointingly, in three independent phase III trials—REMoxTB [10], OFLOTUB [11], RIFAQUIN [12])—the trial goals were not attained and no improvement over the standard of care was observed [13].
(b). Target-based screens
The combination of genomics and bioinformatics had led to new hope for discovering antibacterials in a rational manner and target-based screening was widely embraced by pharmaceutical companies and academia [14]. Having validated that the gene for the chosen target was essential for growth of the bacteria by gene replacement or saturation transposon mutagenesis, high-throughput screens of huge libraries of compounds were implemented against the purified target, usually an enzyme. The inhibitors or ‘hits’ that arose could then be converted into leads by harnessing medicinal chemistry, structural biology and rational drug design. The first steps were generally successful but the process invariably failed when the compounds were tested for antibacterial activity in broth or in infection models. To illustrate this we can cite the example of the essential serine/threonine kinase PknB of M. tuberculosis [15] for which inhibitors with picomolar IC50 values were produced, but none of these showed a minimal inhibitory concentration (MIC) against the pathogen below 100 µg ml−1 (MM4TB unpublished; http://www.mm4tb.org/). There are many reasons to explain this discrepancy and in the case cited it was the inability of the inhibitor to penetrate the cell. Indeed, the thick, complex and hydrophobic cell wall of tubercle bacilli represents a formidable permeability barrier. Other explanations for the disconnect between IC50 and MIC include the target not being required during infection, functional redundancy and the ability of the pathogen to scavenge metabolites from the host cell. Reassuringly, attempts at developing leads for other pathogenic bacteria by this approach were equally disappointing, and most experts regard this era in drug discovery as a marked failure that has directly contributed to the disengagement of pharmaceutical companies from the antibacterial sector [14,16]. In the field of tuberculosis, these disappointments and setbacks triggered the return to a tried and trusted method, phenotypic screening, the very approach that was used to discover the first anti-tubercular agents in the 1950s and 1960s.
(c). Phenotypic or whole-cell screens
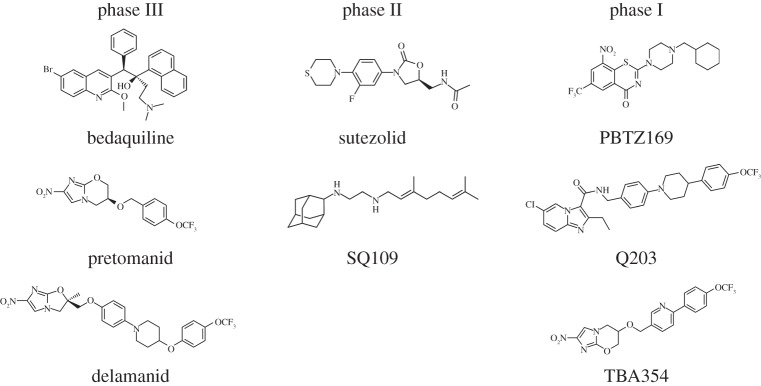
The disappointments of target-based screening led to the realization that pharmacological validation of targets was a more reliable route to lead finding and a better predictor of success against the pathogen (figure 1). Three of the drugs now in phase III clinical trials were found by means of their whole-cell activity and, in chronological order, these were pretomanid [17], bedaquilin [18] and delamanid [19]. Bedaquilin, a diarylquinoline, is particularly interesting as it inhibits the c subunit of ATP-synthase of M. tuberculosis with great specificity, an enzyme that would not have been chosen for target-based screening due to its ubiquity. Pretomanid and delamanid are both nitro-imidazole pro-drugs that require activation by an F420-dependent nitroreductase, Ddn. Their cidal activity likely stems from the pleiotropic action of the nitric oxide thereby released [7]. TBA354, a pretomanid back-up with several advantages, is currently in phase I trials (figure 1).
Figure 1.
Structures of drug candidates in clinical trials.
Other drug candidates that are in earlier stages of clinical testing and were also found by phenotypic screens include the ethylene diamine, SQ109 [20], the imidazopyridine, Q203 [21] and the benzothiazinone, PBTZ169 [22] (figure 1). All three of these candidates inhibit novel protein targets (MmpL3, QcrB, DprE1) in M. tuberculosis that later transpired to be ‘promiscuous’ [23]. Many other hits were uncovered by phenotypic screening but as they have been less intensively investigated I will not discuss them here [24–26].
(d). Promiscuous targets
A puzzling outcome of many phenotypic screens against M. tuberculosis was the finding that the same targets were repeatedly found in many different laboratories despite the use of different compound libraries and assays. Such targets were termed ‘promiscuous’ [23] as they seemed to be affected by a broad range of unrelated pharmacophores (table 1).
Table 1.
Inhibitors of promiscuous targets.
| target | compound class | references |
|---|---|---|
| DprE1 | 1,3-benzothiazin-4-ones (BTZs) (covalent) | [22,27] |
| DprE1 | benzoquinoxaline (covalent) | [28] |
| DprE1 | dinitrobenzamides (covalent) | [29] |
| DprE1 | nitro-substituted triazole (covalent) | [25] |
| DprE1 | benzothiazoles (covalent) | [30] |
| DprE1 | benzothiazole-thiophene (non-covalent) | [31] |
| DprE1 | 2-carboxyquinoxalines (non-covalent) | [32] |
| DprE1 | 1,4-azaindole (non-covalent) | [33] |
| DprE1 | pyrazolopyridone (non-covalent) | [34] |
| MmpL3 | ethylene diamines | [20] |
| MmpL3 | 1,5-diarylpyrrole BM212 | [35] |
| MmpL3 | benzimidazole C215 | [25] |
| MmpL3 | tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamides | [26,36] |
| MmpL3 | N-benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyrans | [36] |
| MmpL3 | indolecarboxamides | [37,38] |
| MmpL3 | adamantyl ureas | [39] |
| MmpL3 | spiropiperidines | [40] |
| Pks13 | thiophenes | [41–43] |
| Pks13 | benzofurans | [26] |
| QcrB | imidazo[1,2-a]pyridine amides (IPA) | [21,44–47] |
| QcrB | imidazo[4,5-c]pyridines | [47] |
| QcrB | piperazine-pyrazole | [47] |
| QcrB | triazolo[3,4-b][1,3,4]thiadiazole | [47] |
| QcrB | oxoquinolines | [47] |
| QcrB | lansoprazole sulphide | [48] |
(i). DprE1
The sole arabinose precursor for synthesis of the critical cell wall components arabinogalactan and lipoarabinomannan is produced by decaprenyl-phosphoryl-β-d-ribose epimerization resulting from two enzymes acting concertedly. First, the flavoprotein DprE1 oxidizes decaprenyl-phosphoryl-β-d-ribose to decaprenyl-phosphoryl-β-d-keto-pento-furanose that is subsequently reduced by the NADH-containing DprE2 to decaprenyl-phosphoryl-β-d-arabinose [49]. Both DprE enzymes are essential [49,50] and, in 2009, DprE1 was discovered to be the target of the 1,3-benzothiazin-4-ones (BTZs), exceptionally potent nitroaromatic inhibitors [27]. The BTZs are suicide inhibitors that undergo nitroreduction by the FADH2-containing form of DprE1 resulting in a nitroso species that covalently binds to a cysteine residue in the active site, thereby irreversibly inactivating the flavoenzyme and thus blocking arabinose production.
Later, several other series of nitroaromatic DprE1 inhibitors were found and, like BTZ, these all form covalent adducts with the active site cysteine [7,49]. These compounds include the dinitrobenzamides, benzoxyquinoxalines, benzothiazoles and nitro-substituted triazoles (table 1). Subsequently, as DprE1 is a highly vulnerable target of M. tuberculosis, several series of non-covalent inhibitors were also developed. Details of the covalent and non-covalent inhibitors may be found in table 1.
(ii). MmpL3
The broadest of the promiscuous targets is the MmpL3 transporter, a member of the resistance-nodulation-division (RND) superfamily (table 1). As MmpL3 belongs to the RND family, there was concern that it might be responsible for efflux-mediated resistance rather than represent a target per se. This concern was largely dispelled when it was demonstrated that treatment with certain compounds, such as SQ109 or the adamantyl urea AU1235, abolishes the ability of MmpL3 to translocate trehalose monomycolate to the periplasm as part of mycolic acid biogenesis. However, as the transmembrane electrochemical proton gradient that energizes this translocation process is dissipated by many of the MmpL3 inhibitors tested, the ability of these compounds to accept protons may explain the ‘promiscuity’ [51]. Furthermore, it is also conceivable that such inhibitors may act pleiotropically and inhibit other enzymes that harness the proton gradient.
(iii). QcrB
Several research groups identified this target independently and most of the inhibitors were based on the imidazopyridine amide scaffold [21,44–46] (table 1). QcrB is the b subunit of cytochrome bc1 oxidase and, consistent with its function in electron transport, inhibition of QcrB leads to ATP depletion. The most advanced candidate, termed Q203, is currently in phase I trials (figure 1) and the initial lead compound was found by high content screening [52]. Laboratory mutants resistant to Q203 all harbour the missense mutation T313A and, based on structural models, this appears to map to the menaquinol binding site of QcrB near to the site of interaction with the Rieske protein (QcrA). More recently, a further four non-imidazopyridine amide scaffolds (table 1) were shown to act as QcrB inhibitors by Arora et al. [47] and a variety of new resistance-conferring mutations were mapped to QcrB, in the vicinity of T313. This may explain why all five scaffolds show extensive cross-resistance in studies with mutants. These investigators caution that M. tuberculosis may be able to overcome QcrB inhibition by using cytochrome bd oxidase as an alternative electron acceptor when oxygen tension changes.
(iv). Pks13
This multi-domain, multifunction polyketide synthase is essential for mycolic acid production. Among the many enzymatic steps catalysed by Pks13 are formation of the α-alkyl-β-ketoester branched-chain precursors by condensation of a C24–C26 long-chain fatty acid with a C40–C60 meromycolate chain. Pks13 then uses its thioesterase activity to release these nascent mycolates and transfers them to trehalose thereby forming the trehalose monomycolate precursor [53]. Several groups have independently found thiophene and benzofuran compounds [26,41,42] that target Pks13, thus ablating mycolic acid synthesis [43]. Ioerger et al. discovered a benzofuran that seemingly inhibits the thioesterase activity (residues 1400–1700) as resistance mutations (D1644G and D1607N) mapped there [26]. By contrast, the site of thiophene inhibition is distinct, as a resistant mutant harbours a F79S substitution that is predicted to be close to the phospho-pantetheine attachment site [43].
(e). Accounting for promiscuity
Goldman noted that membrane protein targets were over-represented in the hits from phenotypic screens and suggested that this might be explained by the relative hydrophobicity of the compounds concerned as their logP-values often exceeded 4 [54]. Another possible explanation may be due to such compounds acting as uncoupling agents that can dissipate the proton motive force required for ATP synthesis [55]. As described in §2d(ii), several MmpL3 inhibitors have this ability. Some of these inhibitors also have a second activity and are able to block the activity of certain key enzymes as exemplified by SQ109 that impacts the 1,4-dihydroxy-2-naphthoate octaprenyltransferase, MenA, required for menaquinone biosynthesis [56].
Another explanation may be provided by the location of the targets themselves. For instance, it has recently been shown that DprE1 is located on the periplasmic side of the plasma membrane, predominantly at the old cell pole [57]. Cell wall biosynthesis is spatially and temporally coordinated, occurring primarily at the poles. The proteins MurG and DivIVA (Wag31) involved in peptidoglycan synthesis; the enzyme GlfT2, that acts downstream of DprE1 and is required for arabinogalactan production; MmpL3, Pks13 and several FAS-II enzymes involved in mycolic acid synthesis and translocation, have all been localized at the cell pole [58,59]. Furthermore, the active site of QcrB is also periplasmic although the precise localization of the enzyme in the plasma membrane has not been defined. Thus, drugs inhibiting these functions do not have to cross the plasma membrane to reach their targets and the corresponding enzymes may be more vulnerable for topological reasons. Indeed, this has led Carel et al. [59] to propose that, during growth, the cell pole may be the Achilles' heel of the tubercle bacillus.
3. Intracellular screens
Given the limitations of both target-based and in vitro phenotypic screens, some investigators proposed that ex vivo screens, employing macrophages infected with M. tuberculosis, would provide a new means of detecting novel inhibitors and discovering targets that might be more relevant during infection. Such cell-based screens offer several major advantages: they provide information about the uptake and intracellular activity of compounds and also enable cytotoxic compounds to be identified early in the screening cascade. Cell-based screens also have the potential to identify host functions that influence intracellular survival of tubercle bacilli and could thus serve as targets for host-directed therapy, although this will not be discussed further here [60]. A particularly elegant implementation of this strategy was provided by high content screening [61], an approach that uses automated confocal fluorescence microscopy to identify compounds that interfere with the replication of tubercle bacilli within macrophages. Such a screen uncovered 135 active compounds with potent intracellular anti-mycobacterial efficacy and no host cell toxicity among a 57 000-strong library of small molecules [29]. These included the dinitrobenzamide derivatives that were shown to be DprE1 inhibitors (table 1) and the hit [21] that later became Q203 (table 1 and figure 1).
In a variation of this screen, an M. tuberculosis strain that constitutively expressed the fluorescent protein mCherry was used to find hits in a collection of approximately 340 000 synthetic compounds [62]. This uncovered two distinct subsets of compounds: those that were universally active, inhibiting mycobacterial growth in vitro and inside macrophages, and those that were conditionally active, inhibiting intracellular but not in vitro growth of M. tuberculosis. Further investigation revealed that some of the latter inhibitors blocked cholesterol catabolism in both settings, thereby limiting growth. Other hit compounds targeted PrpC, the 2-methylcitrate synthase, required for assimilation of propionyl-CoA derived from cholesterol into the TCA cycle, while a further group of hits modulated the adenylate cyclase Rv1625/Cya. The latter finding indicates that cyclic-AMP regulates cholesterol utilization and propionate metabolism in M. tuberculosis [62].
Rybniker et al. [48] devised an innovative intracellular screen to find hits that protected lung fibroblasts from the cytotoxicity associated with M. tuberculosis. A counter-screen against in vitro grown M. tuberculosis enabled known antibiotics to be identified in a collection of US Federal Drug Administration-approved drugs and eliminated from further studies. The proton pump inhibitor lansoprazole (LPZ), an over-the-counter drug used to remedy heartburn and other acid-mediated disorders, was found to be highly active in this screen but only after its intracellular reduction to lansoprazole sulphide (LPZS). Unlike LPZ, LPZS was also active in broth, displaying an MIC of approximately 1 µM, and this enabled the target to be identified after genome sequencing of LPZS-resistant mutants. The same missense mutation (L176P) was found in the qcrB gene of three independent mutants, thus validating cytochrome bc1 oxidase as a bona fide drug target in an infection model. Although the L176P mutation maps to the active site, no significant cross-resistance was observed between LPZS and imidazopyridine amides such as Q203 [48]. LPZS showed modest activity in the acute model of murine tuberculosis and this investigation demonstrates that repurposing drugs developed for other therapeutic areas can be a viable strategy for tuberculosis.
4. Towards anti-virulence drugs
An alternative approach to antibacterial drug discovery that is gaining credence is to identify compounds targeting functions required for pathogenesis or disease. These lead compounds could be developed as anti-virulence drugs for use in conjunction with conventional anti-tubercular agents. Such functions are not required for growth in synthetic media so can only be accessed using ex vivo or in vivo models. Anti-virulence drugs have a number of potential advantages as they should be less prone to resistance than bactericidal drugs, especially to transferable resistance mechanisms. They will also be pathogen-specific, therefore sparing the host's microbiota, and should reduce tissue damage and the resultant pathology [63]. Anti-virulence drugs could then be used in conjunction with conventional anti-tubercular agents.
The ESX-1 type VII protein secretion system is the major virulence determinant of M. tuberculosis and is required for host cell entry, phagosome escape and intercellular spread [64–69]. The best-characterized virulence protein secreted by ESX-1 is EsxA; this helical hairpin protein is endowed with membranolytic and cytolytic activities, and triggers early mechanisms of innate immunity such as the production of type I interferons [70]. It is also a major T-cell antigen and contributes to adaptive immune responses [64,69].
Secretion of EsxA and other ESX-1 substrates requires the activity of at least three different ATPases, EccA1, EccCa1 and EccCb1 [65]. Rybniker et al. [71] therefore exploited a fibroblast survival assay to screen a kinase inhibitor library for compounds that protect against M. tuberculosis-mediated cytotoxicity and reasoned that the nucleoside analogues present therein might inhibit these ATPase activities. The approach was successful, disclosing two series of compounds that prevented secretion of EsxA.
The first series comprised benzothiophene (BTP) derivatives that probably act by inhibiting MprB, the histidine kinase of the MprAB two-component signal transduction system because the lead compound BTP15 was shown to act as a kinase inhibitor in an in vitro MprB autophosphorylation assay. Perturbing the regulatory loop mediated by this signal transduction system leads to downregulation of an operon, espACD, required to produce a critical part of the secretion apparatus [72]. The mechanism of action of the second series, the benzyloxybenzylidine hydrazine (BBH) compounds, was less clear. Exposure of M. tuberculosis to the lead compound, BBH7, disrupted metal ion homeostasis and increased outer membrane permeability. The tubercle bacillus responded by upregulating the ESX-1 ATPases, EccCa1 and EccCb1, but EsxA secretion was blocked by BBH7 by an unknown mechanism. After exposing infected macrophages to BTP15 or BBH7, phagosomes containing mycobacteria fuse with lysosomes, thereby severely inhibiting intracellular growth.
As ESX-1 is a key player in both the innate and adaptive immune responses its inhibition by pharmacological means should open avenues for immunomodulation. Ablasser and her colleagues tested the effect of BTP15 and BBH7 on innate immune responses and found, as predicted from earlier investigations, that downregulation of ESX-1 greatly diminished the production of type I interferons but did not impact the levels of the cytokine IL-1β [73]. Thus, pharmacologically manipulating the ESX-1 secretion system can sway the M. tuberculosis-triggered immune cytokine pattern towards host-protective IL-1β production, by accentuating intracellular sensing of mycobacterial ligands via the inflammasome complex, and also alleviating the type I interferon response that is believed to favour the pathogen.
Ethoxzolamide, a sulphonamide drug used as an anti-diuretic and in glaucoma treatment, is another interesting example of a potential anti-virulence inhibitor. Johnson et al. [74] used an acidic pH-inducible fluorescent reporter strain of M. tuberculosis that is fully dependent on PhoPR regulation to identify compounds that might inhibit this two-component system. PhoPR acts pleiotropically in response to acid stress, and among the genes it regulates are several encoding components of the ESX-1 system [75,76]. Ethoxzolamide inhibits human carbonic anhydrase and displays the same activity against M. tuberculosis, which has three such enzymes, as well as downregulating ESX-1 secretion. Exposure to ethoxzolamide protects both macrophages and mice against M. tuberculosis, probably by indirectly downregulating expression of the ESX-1 system, although another important virulence factor, the lipid phthiocerol dimycocerosate, is also regulated by PhoPR. This discovery was interesting for two reasons: firstly, it identifies carbonic anhydrase activity as a likely cue for PhoPR; secondly, it provides an opportunity to repurpose ethoxzolamide, or a related molecule, for tuberculosis therapy.
5. Conclusion and perspectives
Significant strides have been made in reducing the mortality resulting from tuberculosis as the death rate has declined by over 40% since 2000 to around 1.5 million/year today. This reduction was owing largely to improved operational procedures and better access to treatment resulting from increased funding contributed by the Global Fund to fight AIDS, Tuberculosis and Malaria, the President's Emergency Plan for AIDS Relief (PEPFAR) and other such bodies. However, during the same period the number of new cases of tuberculosis has remained relatively constant and shows few signs of receding, with drug resistance continuing to climb. It is frequently proposed that vaccination is the only intervention likely to make an impact on the incidence of the disease, but numerous attempts to develop vaccines that are more efficacious in humans than the BCG (Bacille de Calmette et Guérin) vaccine have failed despite the repeated efforts of many talented scientists [77].
In this article, I have discussed the obstacles in finding lead compounds and summarized some of the scientific achievements that have undoubtedly been made in the field of tuberculosis drug discovery and development, thanks to the efforts of the global research community and their funding agencies. However, the likelihood of any of this research being translated into drugs that reach the clinic is steadily diminishing. On the one hand, this is due to the dwindling number of pharmaceutical companies still engaged in this area—it should be recalled that AstraZeneca, Novartis and Pfizer have all closed their tuberculosis research and development (R&D) programmes in recent years—and on the other to the chronic lack of funds needed to support such initiatives. In the year 2000, Romano Prodi, the President of the European Commission, declared ‘I want to see the EU playing a larger and more effective role in assisting developing countries to confront these epidemics'. He would be disappointed to learn that in its Horizon 2020 programme, the European Commission has earmarked no funds for R&D on tuberculosis drugs and diagnostics. This is especially ironic at a time when the European Union proposes to host three million refugees from the Middle East, many of whom will soon present with tuberculosis, as substantiated by the findings of a recent survey of 488 unaccompanied Syrian minors in Berlin; 11% presented with active tuberculosis [78]. Action is required!
Acknowledgements
Many thank to Drs Jeffrey Chen, Gyorgy Kéri, Vadim Makarov and Jan Rybniker for helpful discussions and advice.
Competing interests
I am a named inventor on patents pertaining to work discussed in this article.
Funding
I thank the European Commission's Seventh Framework Programme (grant no. 260872) and the Swiss National Science Foundation (31003A-162641) for supporting my work.
References
- 1.Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. Oxford, UK: Oxford University Press. [Google Scholar]
- 2.Lewis K. 2012. Antibiotics: recover the lost art of drug discovery. Nature 485, 439–440. ( 10.1038/485439a) [DOI] [PubMed] [Google Scholar]
- 3.Cole ST. 2014. Who will develop new antibacterial agents? Phil. Trans. R. Soc. B 369, 20130430 ( 10.1098/rstb.2013.0430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2015. Global tuberculosis report 2015. Geneva, Switzerland: World Health Organization.
- 5.Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. ( 10.1038/31159) [DOI] [PubMed] [Google Scholar]
- 6.Diacon AH, et al. 2012. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet 380, 986–993. ( 10.1016/S0140-6736(12)61080-0) [DOI] [PubMed] [Google Scholar]
- 7.Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 12, 388–404. ( 10.1038/nrd4001) [DOI] [PubMed] [Google Scholar]
- 8.Nuermberger EL, Yoshimatsu T, Tyagi S, O'Brien RJ, Vernon AN, Chaisson RE, Bishai WR, Grosset JH. 2004. Moxifloxacin-containing regimen greatly reduces time to culture conversion in murine tuberculosis. Am. J. Respir. Crit. Care Med. 169, 421–426. ( 10.1164/rccm.200310-1380OC) [DOI] [PubMed] [Google Scholar]
- 9.Rustomjee R, et al. 2008. A phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 12, 128–138. [PubMed] [Google Scholar]
- 10.Gillespie SH, et al. 2014. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N. Engl. J. Med. 371, 1577–1587. ( 10.1056/NEJMoa1407426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merle CS, et al. 2014. A four-month gatifloxacin-containing regimen for treating tuberculosis. N. Engl. J. Med. 371, 1588–1598. ( 10.1056/NEJMoa1315817) [DOI] [PubMed] [Google Scholar]
- 12.Jindani A, et al. 2014. High-dose rifapentine with moxifloxacin for pulmonary tuberculosis. N. Engl. J. Med. 371, 1599–1608. ( 10.1056/NEJMoa1314210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner DF, Mizrahi V. 2014. Shortening treatment for tuberculosis--to basics. N. Engl. J. Med. 371, 1642–1643. ( 10.1056/NEJMe1410977) [DOI] [PubMed] [Google Scholar]
- 14.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40. ( 10.1038/nrd2201) [DOI] [PubMed] [Google Scholar]
- 15.Fernandez P, Saint-Joanis B, Barilone N, Jackson M, Gicquel B, Cole ST, Alzari PM. 2006. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188, 7778–7784. ( 10.1128/JB.00963-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sams-Dodd F. 2005. Target-based drug discovery: is something wrong? Drug Discov. Today 10, 139–147. ( 10.1016/S1359-6446(04)03316-1) [DOI] [PubMed] [Google Scholar]
- 17.Stover CK, et al. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405, 962–966. ( 10.1038/35016103) [DOI] [PubMed] [Google Scholar]
- 18.Andries K, et al. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227. ( 10.1126/science.1106753) [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. 2006. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 3, e466 ( 10.1371/journal.pmed.0030466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacksteder KA, Protopopova M, Barry CE 3rd, Andries K, Nacy CA. 2012. Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiol. 7, 823–837. ( 10.2217/fmb.12.56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pethe K, et al. 2013. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 19, 1157–1160. ( 10.1038/nm.3262) [DOI] [PubMed] [Google Scholar]
- 22.Makarov V, et al. 2014. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol. Med. 6, 372–383. ( 10.1002/emmm.201303575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechartier B, Rybniker J, Zumla A, Cole ST. 2014. Tuberculosis drug discovery in the post-post-genomic era. EMBO Mol. Med. 6, 158–168. ( 10.1002/emmm.201201772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballell L, et al. 2013. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem 8, 313–321. ( 10.1002/cmdc.201200428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley SA, et al. 2012. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem. Biol. 7, 1377–1384. ( 10.1021/cb300151m) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioerger TR, et al. 2013. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS ONE 8, e75245 ( 10.1371/journal.pone.0075245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarov V, et al. 2009. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324, 801–804. ( 10.1126/science.1171583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnet S, et al. 2010. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis 90, 354–360. ( 10.1016/j.tube.2010.09.001) [DOI] [PubMed] [Google Scholar]
- 29.Christophe T, et al. 2009. High content screening identifies decaprenyl-phosphoribose 2' epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog. 5, e1000645 ( 10.1371/journal.ppat.1000645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landge S, et al. 2015. Discovery of benzothiazoles as antimycobacterial agents: synthesis, structure-activity relationships and binding studies with Mycobacterium tuberculosis decaprenylphosphoryl-β-d-ribose 2'-oxidase. Bioorg. Med. Chem. 23, 7694–7710. ( 10.1016/j.bmc.2015.11.017) [DOI] [PubMed] [Google Scholar]
- 31.Wang F, et al. 2013. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc. Natl Acad. Sci. USA 110, E2510–E2517. ( 10.1073/pnas.1309171110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neres J, et al. 2015. 2-Carboxyquinoxalines kill Mycobacterium tuberculosis through noncovalent inhibition of DprE1. ACS Chem. Biol. 10, 705–714. ( 10.1021/cb5007163) [DOI] [PubMed] [Google Scholar]
- 33.Chatterji M, et al. 2014. 1,4-Azaindole, a potential drug candidate for treatment of tuberculosis. Antimicrob. Agents Chemother. 58, 5325–5331. ( 10.1128/AAC.03233-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panda M, et al. 2014. Discovery of pyrazolopyridones as a novel class of noncovalent DprE1 inhibitor with potent anti-mycobacterial activity. J. Med. Chem. 57, 4761–4771. ( 10.1021/jm5002937) [DOI] [PubMed] [Google Scholar]
- 35.La Rosa V, et al. 2012. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324–331. ( 10.1128/AAC.05270-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remuinan MJ, et al. 2013. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6',7'-dihydrospiro[piperidine-4,4'-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS ONE 8, e60933 ( 10.1371/journal.pone.0060933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao SP, et al. 2013. Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci. Transl. Med. 5, 214ra168. ( 10.1126/scitranslmed.3007355) [DOI] [PubMed] [Google Scholar]
- 38.Lun S, et al. 2013. Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat. Commun. 4, 2907 ( 10.1038/ncomms3907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grzegorzewicz AE, et al. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341. ( 10.1038/nchembio.794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tantry SJ, et al. 2015. Whole cell screen based identification of spiropiperidines with potent antitubercular properties. Bioorg. Med. Chem. Lett. 25, 3234–3245. ( 10.1016/j.bmcl.2015.05.087) [DOI] [PubMed] [Google Scholar]
- 41.Maddry JA, et al. 2009. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis 89, 354–363. ( 10.1016/j.tube.2009.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ananthan S, et al. 2009. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis 89, 334–353. ( 10.1016/j.tube.2009.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson R, et al. 2013. Antituberculosis thiophenes define a requirement for Pks13 in mycolic acid biosynthesis. Nat. Chem. Biol. 9, 499–506. ( 10.1038/nchembio.1277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrahams KA, et al. 2012. Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PLoS ONE 7, e52951 ( 10.1371/journal.pone.0052951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moraski GC, et al. 2013. Advancement of imidazo[1,2-]pyridines with improved pharmacokinetics and nanomolar activity against. ACS Med. Chem. Lett. 4, 675–679. ( 10.1021/ml400088y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khoje AD, Charnock C, Wan B, Franzblau S, Gundersen LL. 2011. Synthesis and antimycobacterial activities of non-purine analogs of 6-aryl-9-benzylpurines: imidazopyridines, pyrrolopyridines, benzimidazoles, and indoles. Bioorg. Med. Chem. 19, 3483–3491. ( 10.1016/j.bmc.2011.04.023) [DOI] [PubMed] [Google Scholar]
- 47.Arora K, et al. 2014. Respiratory flexibility in response to inhibition of cytochrome c oxidase in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6962–6965. ( 10.1128/AAC.03486-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rybniker J, Vocat A, Sala C, Busso P, Pojer F, Benjak A, Cole ST. 2015. Lansoprazole is an antituberculous prodrug targeting cytochrome bc1. Nat. Commun. 6, 7659 ( 10.1038/ncomms8659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mikusova K, Makarov V, Neres J. 2014. DprE1--from the discovery to the promising tuberculosis drug target. Curr. Pharmaceut. Des. 20, 4379–4403. ( 10.2174/138161282027140630122724) [DOI] [PubMed] [Google Scholar]
- 50.Kolly GS, et al. 2014. Assessing the essentiality of the decaprenyl-phospho-d-arabinofuranose pathway in Mycobacterium tuberculosis using conditional mutants. Mol. Microbiol. 92, 194–211. ( 10.1111/mmi.12546) [DOI] [PubMed] [Google Scholar]
- 51.Li W, et al. 2014. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6413–6423. ( 10.1128/AAC.03229-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brodin P, Christophe T. 2011. High-content screening in infectious diseases. Curr. Opin. Chem. Biol. 15, 534–539. ( 10.1016/j.cbpa.2011.05.023) [DOI] [PubMed] [Google Scholar]
- 53.Gavalda S, et al. 2014. The polyketide synthase Pks13 catalyzes a novel mechanism of lipid transfer in mycobacteria. Chem. Biol. 21, 1660–1669. ( 10.1016/j.chembiol.2014.10.011) [DOI] [PubMed] [Google Scholar]
- 54.Goldman RC. 2013. Why are membrane targets discovered by phenotypic screens and genome sequencing in Mycobacterium tuberculosis? Tuberculosis 93, 569–588. ( 10.1016/j.tube.2013.09.003) [DOI] [PubMed] [Google Scholar]
- 55.Feng X, et al. 2015. Antiinfectives targeting enzymes and the proton motive force. Proc. Natl Acad. Sci. USA 112, E7073–E7082. ( 10.1073/pnas.1521988112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li K, et al. 2014. Multitarget drug discovery for tuberculosis and other infectious diseases. J. Med. Chem. 57, 3126–3139. ( 10.1021/jm500131s) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brecik M, et al. 2015. DprE1 is a vulnerable tuberculosis drug target due to its cell wall localization. ACS Chem. Biol. 10, 1631–1636. ( 10.1021/acschembio.5b00237) [DOI] [PubMed] [Google Scholar]
- 58.Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. 2014. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc. Natl Acad. Sci. USA 111, E3243–E3251. ( 10.1073/pnas.1402158111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carel C, Nukdee K, Cantaloube S, Bonne M, Diagne CT, Laval F, Daffe M, Zerbib D. 2014. Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum. PLoS ONE 9, e97148 ( 10.1371/journal.pone.0097148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zumla A, et al. 2015. Towards host-directed therapies for tuberculosis. Nat. Rev. Drug Discov. 14, 511–512. ( 10.1038/nrd4696) [DOI] [PubMed] [Google Scholar]
- 61.Christophe T, Ewann F, Jeon HK, Cechetto J, Brodin P. 2010. High-content imaging of Mycobacterium tuberculosis-infected macrophages: an in vitro model for tuberculosis drug discovery. Future Med. Chem. 2, 1283–1293. ( 10.4155/fmc.10.223) [DOI] [PubMed] [Google Scholar]
- 62.VanderVen BC, et al. 2015. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment. PLoS Pathog. 11, e1004679 ( 10.1371/journal.ppat.1004679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JM, Pojer F, Blasco B, Cole ST. 2010. Towards anti-virulence drugs targeting ESX-1 mediated pathogenesis of Mycobacterium tuberculosis. Drug Discov. Today 7, e25–e31. ( 10.1016/j.ddmec.2010.09.002) [DOI] [Google Scholar]
- 64.Bitter W, et al. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5, e1000507 ( 10.1371/journal.ppat.1000507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Houben EN, Korotkov KV, Bitter W. 2014. Take five—type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta 1843, 1707–1716. ( 10.1016/j.bbamcr.2013.11.003) [DOI] [PubMed] [Google Scholar]
- 66.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8, e1002507 ( 10.1371/journal.ppat.1002507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simeone R, Bottai D, Brosch R. 2009. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 12, 4–10. ( 10.1016/j.mib.2008.11.003) [DOI] [PubMed] [Google Scholar]
- 68.Simeone R, Bottai D, Frigui W, Majlessi L, Brosch R. 2015. ESX/type VII secretion systems of mycobacteria: Insights into evolution, pathogenicity and protection. Tuberculosis 95(Suppl 1), S150–S154. ( 10.1016/j.tube.2015.02.019) [DOI] [PubMed] [Google Scholar]
- 69.Simeone R, Sayes F, Song O, Groschel MI, Brodin P, Brosch R, Majlessi L. 2015. Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 11, e1004650 ( 10.1371/journal.ppat.1004650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. 2007. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178, 3143–3152. ( 10.4049/jimmunol.178.5.3143) [DOI] [PubMed] [Google Scholar]
- 71.Rybniker J, et al. 2014. Anticytolytic screen identifies inhibitors of mycobacterial virulence protein secretion. Cell Host Microbe 16, 538–548. ( 10.1016/j.chom.2014.09.008) [DOI] [PubMed] [Google Scholar]
- 72.Pang X, Samten B, Cao G, Wang X, Tvinnereim AR, Chen XL, Howard ST. 2013. MprAB regulates the espA operon in Mycobacterium tuberculosis and modulates ESX-1 function and host cytokine response. J. Bacteriol. 195, 66–75. ( 10.1128/JB.01067-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wassermann R, et al. 2015. Mycobacterium tuberculosis differentially activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe 17, 799–810. ( 10.1016/j.chom.2015.05.003) [DOI] [PubMed] [Google Scholar]
- 74.Johnson BK, Colvin CJ, Needle DB, Mba Medie F, Champion PA, Abramovitch RB. 2015. The carbonic anhydrase inhibitor ethoxzolamide inhibits the Mycobacterium tuberculosis PhoPR regulon and Esx-1 secretion and attenuates virulence. Antimicrob. Agents Chemother. 59, 4436–4445. ( 10.1128/AAC.00719-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernandez-Pando R, Thole J, Behr M, Gicquel B, Martin C. 2008. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS ONE 3, e3496 ( 10.1371/journal.pone.0003496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frigui W, et al. 2008. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 4, e33 ( 10.1371/journal.ppat.0040033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tameris MD, et al. 2013. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 381, 1021–1028. ( 10.1016/S0140-6736(13)60177-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mockenhaupt FP, et al. 2016. Profile of illness in Syrian refugees: a GeoSentinel analysis, 2013 to 2015. Euro Surveill. 21, 30160 ( 10.2807/1560-7917.ES.2016.21.10.30160) [DOI] [PubMed] [Google Scholar]