Fig. 2.
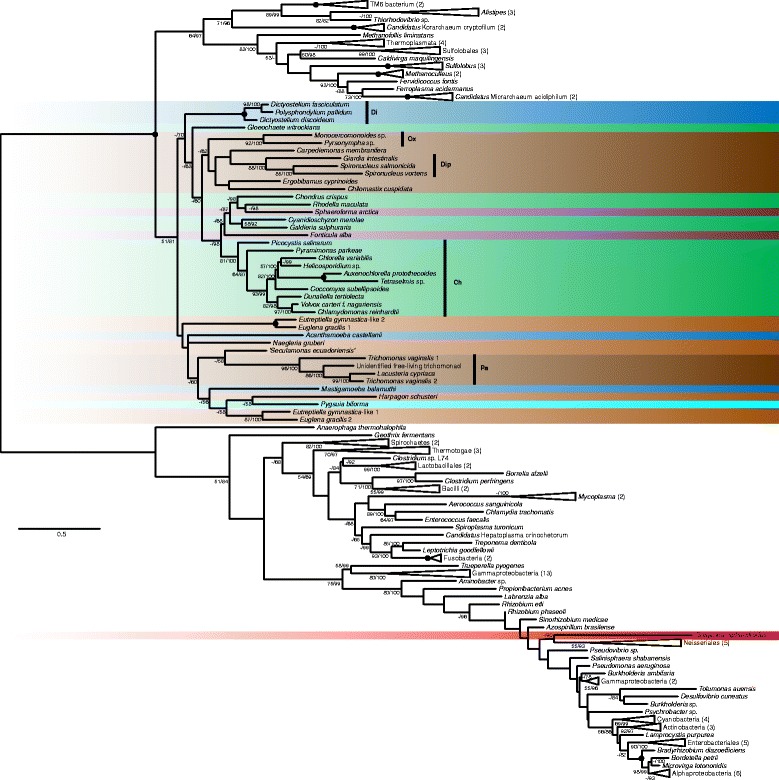
Phylogenetic tree of ADI sequences. The tree based on a 257 positions long protein alignment of 152 sequences was constructed in RAxML using the LG4X + Γ model of substitution. Eukaryotic taxa are highlighted in different colors according to the major group they belong to. The color code is the same as in Fig. 1. The values at nodes represent RAxML bootstrap support/IQ-TREE bootstrap support. Only values above 50 % are shown. Black circles indicate support of 100 %/100 %. Vertical black bars indicate well-supported eukaryotic clades: Pa – Parabasalia; Di – Dictyosteliida; Ch – Chlorophyta; Ox – Oxymonadida; Dip – Diplomonadida. Species with multiple sequences included: Euglena gracilis 1 – GI 109790819; Euglena gracilis 2 – GI 109784514; Eutreptiella gymnastica-like 1 – CAMPEP 0200414012; Eutreptiella gymnastica-like 2 – CAMPEP 0200422928; Trichomonas vaginalis 1 – TVAG 183850; Trichomonas vaginalis 2 – TVAG 344520. The tree is unrooted
