Fig. 8.
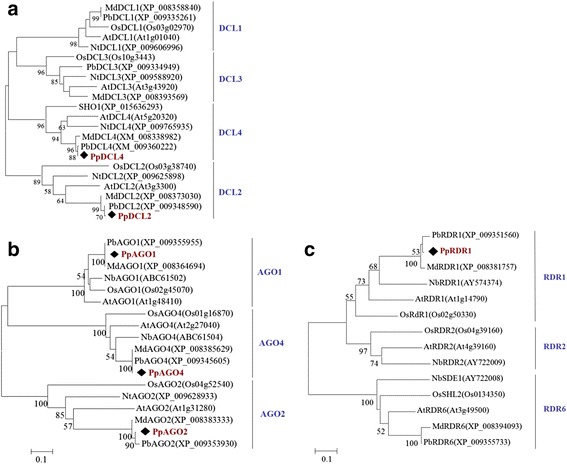
Phylogenetic relationships between the PpDCL2,4, PpAGO1,2,4, and PpRDR1 proteins of P. pyrifolia with their homologues in Pyrus bretschneideri, Malus, Oryza sativa, Nicotiana, and Arabidopsis species. a, b, and c are unrooted Neighbor-joining trees constructed with predicted protein sequences for Dicer-like, Argonaute, and RNA-dependent RNA polymerases from this study and homologous sequences from other isolates deposited into GenBank (gene name followed by GenBank accession number). Statistical analysis was performed with 1,000 bootstrap replicates, and bootstrap values >50 % are shown at branch nodes. Bars represent 0.1 substitutions per site. The predicted P. pyrifolia DCL, AGO, and RDR proteins are shown in red in each group. The abbreviations for the AGO, DCL, and RDR proteins used in the phylogenetic trees are as the follows: Pb, Pyrus bretschneideri; Pp, Pyrus pyrifolia; Md, Malus domestic; Os, Oryza sativa; Nb, Nicotiana benthamiana; Nt, Nicotiana thaliana; At, Arabidopsis thaliana
