Figure 1.
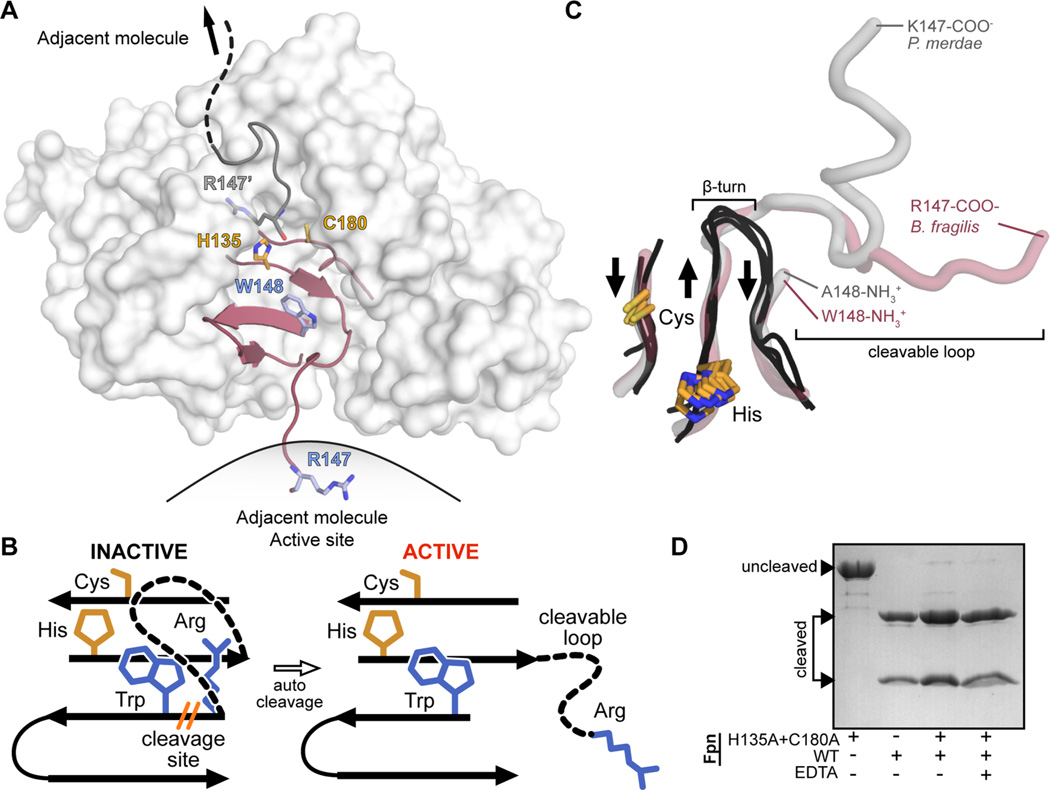
B. fragilis Fpn active site and cleavable activation loop. Residue numbering includes the 19-amino acid predicted signal peptide present at the N-terminus of Fpn. (A) Surface representation of Fpn (white) with the β-strands constituting the active site colored maroon (PDB entry 5DYN). B. fragilis Fpn was crystallized in a cleaved form (cleavage site R147/W148, side chains colored light blue, catalytic dyad H135 and C180, side chains colored orange), with the cleaved region trapped in the active site of the neighboring molecule. (B) Cartoon representation of the B. fragilis Fpn active site and cleavable loop. When Fpn is inactive, the cleavable loop is intact and inhibits the active site composed of the catalytic dyad, H135 and C180 (orange sticks). Fpn is activated upon cis or trans loop cleavage between residues R147 and W148 (blue sticks), which modifies active site accessibility. (C) Superposition of the active site regions of legumain (PDB entry 4AW9), caspase-3 (PDB entry 1NMS), gingipain (PDB entry 4RBM), PmC11 (PDB entry 3UWS), and Fpn structures (PDB entry 5DYN). β-Strands composing the active sites of legumain, caspase-3, and gingipain are shown as black loops, and the equivalent regions in PmC11 and Fpn are show as transparent gray and red tubes, respectively. The aligned catalytic dyad side chains of cysteine and histidine are shown as orange sticks. The β-turn present in legumain, caspase-3, and gingipain structures and the cleaved loops present at the equivalent position in PmC11 and Fpn structures are highlighted. Residues forming the termini of the cleavage sites are annotated. (D) Wild-type Fpn trans cleavage assay, in which Fpn was incubated with the active site mutant FpnH135A+C180A, in the presence (+) or absence (−) of EDTA.