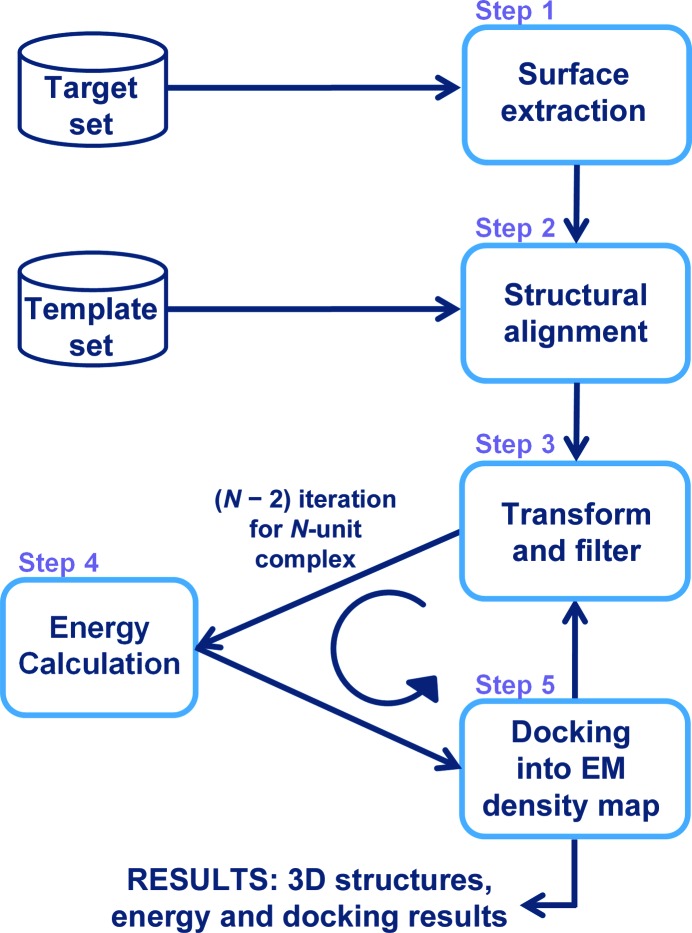
Figure 1.
Flowchart of assembly construction using a 3DEM density map. Step 1: the surfaces of the target proteins are extracted. Step 2: the surfaces of the target proteins are aligned onto the template interfaces of the known interactions. Step 3: target proteins are transformed next to each other with regard to the template interfaces they match; candidate interactions are filtered with regard to hot-spot matches and clashes. Step 4: flexible refinement and energy calculation are performed. Interactions with favourable energies pass this step. Step 5: structures are docked into the 3DEM density map and checked whether they fit. N − 2 iterations are performed through steps 3–5 to construct N-subunit assemblies. The end result consists of assembly structures together with their energies and docked 3DEM density maps. PRISM predicts binary interaction through steps 1–4. It gives the structures of the interactions and their energies as the result.