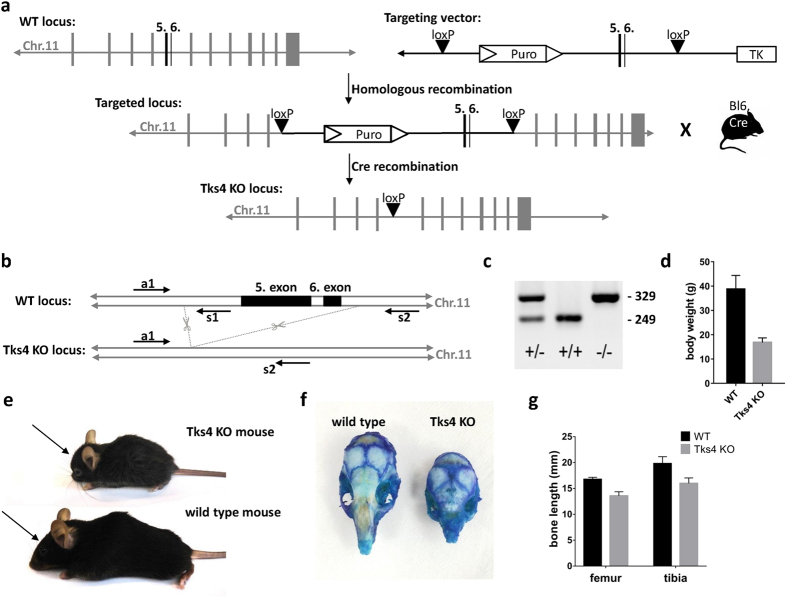
Figure 1. Generation and characterization of Tks4-deficient mice.
(a) Gene targeting strategy to knock out exons 5 and 6 of Tks4. In the targeting vector, exons 5 and 6 were flanked by loxP sites. A puromycin (Puro) resistance gene cassette was inserted into intron 4 and the thymidine kinase gene (TK) was inserted downstream of exon 6 for positive and negative selection, respectively. Mice carrying the mutant floxed allele were crossed with transgenic C57Bl/6 mouse carrying Cre recombinase. (b) Position of deleted exons 5 and 6 are depicted in chromosome 11. The primer set (a1, s1, s2) and the amplified regions (WT: 249 bp, KO: 329 bp) are indicated on the SH3PXD2B wild type (WT) and knock-out (KO) gene. (c) PCR genotyping of heterozygous (+/−), wild type (+/+) and homozygous Tks4 knock-out (−/−) mice. Genomic DNAs obtained from offspring of heterozygous (+/−) mice, were amplified using primer sequences (a1, s1, s2) located near the deleted region. (d) Body weights of 8–10 months old Tks4−/− mice (n = 4) and wild type mice (n = 3). (e) Tks4−/− mouse and wild type littermate. Arrows show the shorter nasal bone of Tks4−/− mouse compared to wild type. (f) Calvarias from an 8 months old wild type and a littermate Tks4−/− mouse were stained with methylene blue. (g) Bone length measurements of 8–12 months old Tks4−/− mice (n = 5) and wild type mice (n = 5). *p < 0.05. An unpaired t-test was used to determine the significance of the difference between means of two groups. Error bars represent s.d.