While chronic lymphocytic leukemia (CLL) is characterized by a strong familial risk, the genetic basis of inherited susceptibility to CLL is largely unknown. The increased risk of Hodgkin Lymphoma (HL) and non-Hodgkin Lymphoma (NHL) in relatives of CLL patients suggests a common etiology to B-cell lymphoproliferative disorders (LPDs) through HLA variation(1). Moreover, since B-cell proliferation is part of an adaptive immune response which can be initiated by major histocompatibility complex (MHC)-restricted T-cell activation, a possible influence of HLA on CLL pathogenesis is plausible.
It has recently been demonstrated that single nucleotide polymorphism (SNP) variation within the 6p21 region can accurately predict alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci(2). Furthermore, HLA alleles can be accurately predicted from the SNPs used in a genome-wide association study (GWAS). To investigate the role of genetic variation in the MHC region in the etiology of CLL we have applied this methodology to SNP data from a GWAS of CLL.
The 517 CLL cases (364 male) analyzed in the GWAS have been previously documented(3). Briefly, they comprised 155 CLL cases with a relative affected with CLL or a related B-cell LPD ascertained through the International CLL linkage consortium (ICLLLC) and 362 cases ascertained through the Leukaemia Research CLL4 trial. The genome-wide SNP scan was conducted using Illumina Infinium HD Human370 Duo BeadChips(3). For controls we made use of genotype data generated by the Wellcome Trust Case Control Consortium on 2,930 individuals from the British 1958 Birth Cohort (58BC). Collection of samples and information from subjects was undertaken with informed consent and ethical review board approval. After imposing rigorous quality control in terms of excluding samples and SNPs with poor call rates and SNPs showing significant departure from Hardy-Weinberg equilibrium, samples showing evidence of relatedness and ancestral differences(3), SNP genotypes were available on 503 cases and 2,698 controls.
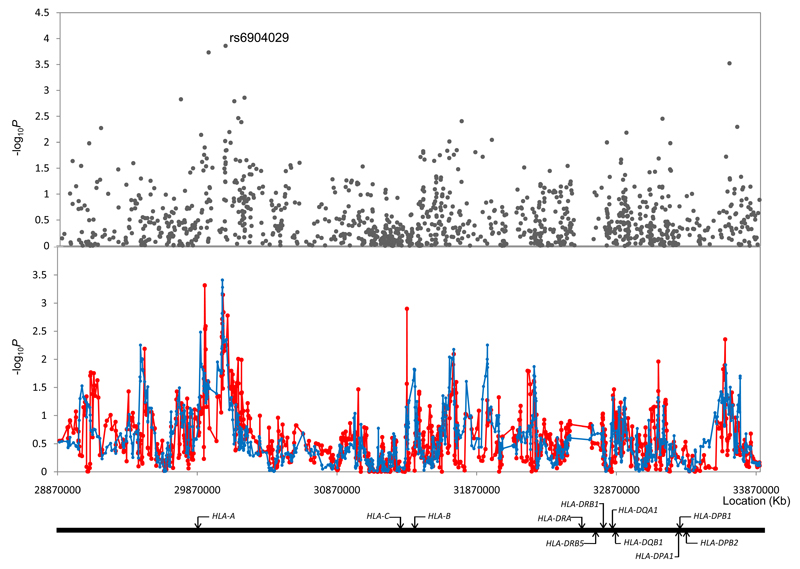
For single SNP and haplotype analysis we considered the MHC at 6p21 to be defined by a 5Mb region bordered by the RFP and MLN genes (rs4324798 at 28,884,096bps and rs767896 at 33,899,493bps; Figure 1). We initially considered the 1,149 SNPs mapping to this 5Mb region analyzing the association between SNP and CLL using the Cochran-Armitage trend test. Odds ratios (ORs) and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression. At the 0.05 threshold 88 SNPs showed evidence of an association compared with an expected of 57.5 (P<0.001). Associations at the 0.01 threshold are reported in Supplementary Table 1. The strongest single association was shown by rs6904029 which maps at 30,051,046bps localizing to the genomic sequence for the non-coding sequence for HLA complex group 9 which is intronic to HLA-A (P=1.38x10-4, Figure 1, Supplementary Table 1). To interrogate the relationship between MHC variation and CLL further we studied haplotypes using HelixTree software v6.4.2 (Golden Helix, Bozeman, MT, USA), using sliding window sizes of 5 and 13 contiguous markers. This analysis provided little evidence for enigmatic disease alleles present on rare haplotypes missed by single SNP analyses (Figure 1).
Figure 1. Association between SNPs and haplotypes mapping to 6p21 and CLL risk.
The x-axis represents the position of each SNP, and the y-axis depicts P values on a minus logarithmic scale. Cochran-Armitage trend test statistics are shown in grey for directly genotyped SNPs in the top panel. Lines in the bottom panel correspond to haplotype test statistics: blue defined by 5 SNPs and red by 13 SNPs. Relative positions of the major HLA genes are also shown. Chromosomal coordinates derived from the National Center for Biotechnology Information, build 36.
To validate the rs6904029 association, using allele-specific PCR KASPar chemistry (KBiosciences), we genotyped an additional series of 919 unrelated CLL cases (599 male) ascertained through the ICLLLC and 1,477 healthy individuals recruited from the National Study of Colorectal Cancer (NSCCG; 1999–2006; n=1,269) and the Royal Marsden Hospital Trust/Institute of Cancer Research Family History and DNA Registry (1999–2004; n=208). Successful genotyping was obtained for 897 cases and 1,429 controls. This analysis provided further evidence for an association between rs6904029 and CLL risk (P=0.01; Table 1) and in the combined analysis the OR for CLL associated with rs6904029-A genotype was 1.24 (95% CI: 1.13-1.36; P=1.00x10-5). The association remained statistically significant after applying a Bonferroni correction to adjust for multiple testing (Padjusted=0.011).
Table 1. Association between rs6904029 and risk of chronic lymphocytic leukemia.
| GWAS | Replication | Combined* | ||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Cases | Controls | ORa (95% CIb) | Cases | Controls | OR (95% CI) | OR (95% CI) | Phet (I2) |
| AA | 67 | 213 | 1.94 (1.40-2.66) | 100 | 113 | 1.52 (1.12-2.07) | 1.71 (1.37-2.13) | 0.28 (13.7%) |
| AG | 212 | 1,109 | 1.18 (0.96-1.45) | 389 | 613 | 1.09 (0.91-1.31) | 1.13 (0.99-1.29) | 0.57 (0.0%) |
| GG | 223 | 1,375 | - | 408 | 703 | - | - | - |
| Total | 502 | 2,697 | 1.32 (1.14-1.52) per allele Ptrend=1.38x10-4 |
897 | 1,429 | 1.18 (1.04-1.34) per allele Ptrend=0.01 |
1.24 (1.13-1.36) Ptrend=1.00x10-5 |
0.25 (25.5%) |
Odds Ratio.
95% Confidence Interval.
Meta-analysis under fixed effects model. Phet, P-value for heterogeneity between studies determined from Cochran’s Q statistic. I2, proportion of the total variation due to heterogeneity calculated; I2 values ≥75% are considered characteristic of large heterogeneity.
To survey the relationship between HLA alleles and CLL risk, we predicted class I and II HLA alleles using HLA*IMP software (version 1.3) making use of a reference database from the HapMap Project and 58BC controls(2). To ensure prediction accuracy, we only analysed alleles that were predicted with a posterior probability >90%, and this criterion was met by >88% of the data. Five alleles showed an association with CLL risk at the 0.05 threshold: HLA-A*0201, HLA-A*3101, HLA-B*1401, HLA-C*0802, HLA-DRB*1101 (Supplementary Table 2). Only the HLA-A*0201 association (P=3.12x10-4) remaining statistically significant after adjustment for multiple testing. Carrier status for HLA-A*0201 conferred a 1.32-fold increased risk of CLL (95% CI: 1.13-1.53). rs6904029 is a proxy for HLA-A*0201 (P<10-7; D’=0.96, r2=0.91) but is not significantly correlated with other HLA-A alleles.
A recent GWAS of CLL reported an association with SNPs mapping to HLA-DRB5 and HLA-DQA1 loci(4). In our study the best evidence for an association at this region was provided by rs660895 which maps to 32,685,358bps (P=0.09) and HLA-DRB*1101 (P=0.038, Supplementary Table 2).
To examine if carrier status for HLA-A*0201 was associated with a restricted immunoglobulin gene usage we made use of previously generated data on CLL4 cases(5). While immunoglobulin heavy chain variable region (IGHV) usage was non-random, with VH3, VH1, and VH4 families expressed at the highest frequencies (50%, 27%, and 15% respectively, Supplementary Table 3), globally there was no difference in usage of specific VH subtypes by HLA-A*0201 genotype in the cases after correction for multiple testing.
Our data provides compelling evidence for a HLA-class I association and specifically that HLA-A*0201 is associated with an increased CLL risk. There is increasing evidence that T-cell dysfunction in CLL may contribute to disease etiology. Specifically, T-cells in CLL may be unable to start, maintain and complete an immune response to the malignant B-cell and other antigens and are involved directly in sustaining the tumor. In addition, in the context of T cell cross-talk, CD4+ T-cells in CLL have been identified in the pseudofollicle/proliferation centers on the tissues involved and their physical contact with CLL cells suggests an important role in the activation and survival of CLL cells(6). A role for the HLA-A*02 allele in evoking an effective immune response is supported by the observation that HLA-A*02 is associated with reduced persistence of hepatitis B viral infection(7) and the finding of an under-representation of HLA-A*02 in patients with tuberculosis(8). The HLA-A*02 allele has also been consistently shown to afford protection against multiple sclerosis(9). HL displays a strong HLA class I association, with under-representation of HLA-A*02 associated with Epstein-Barr virus (EBV)-positive disease (10). Intriguingly in a recent GWAS of HL in the MHC region rs6904029 provided one of the strongest SNP associations for EBV-positive HL, the A-allele conferring an OR of 0.46(11). Following EBV infection, infected memory B-cells escape immune detection by down-regulation of viral antigens. Activation of replicative (i.e. lytic) infection and outgrowth of latently infected cells is kept under tight control by HLA and cytotoxic T-lymphocytes (CTLs). HLA-A*02 and HLA-A*0201 in particular are known to present peptides from a wide range of EBV lytic and latent antigens, including the latent membrane proteins LMP2 and LMP1, hence HLA-A*0201-restricted CTL response provides an eminently biological plausible basis for disease etiology in EBV positive-HL. The CLL association with HLA-A*0201 is however analogous to that shown in nasopharyngeal carcinoma (NPC), whereby an increased risk of NPC is associated with HLA-A*0201 carrier status(12). The non-random usage of variable domain elements of IGHV provides evidence of selection by chronic antigen stimulation or selection through the B-cell receptor. The absence of a strong association between specific HLA-A*0201 genotype and IGHV subtype in CLL cases however argues against a simple environmental basis for disease development.
While we observed a strong class I association with CLL in this study it is entirely plausible, given the over-representation of associations seen at the 0.05 threshold, that genetic variation in other HLA alleles may impact on disease, albeit less profoundly. The recent GWAS of CLL reported by Slager and co-workers found an association between SNPs mapping to the 6p21.32 region which encompasses the HLA-DQA1 and HLA-DRB5 genes(4). It is intriguing that the association was confined to familial CLL cases as familial disease is essentially indistinguishable from CLL(5). The region has been recently identified to harbor variants associated with other B-cell malignancies, including follicular lymphoma and diffuse large B-cell lymphoma(13, 14). Since familial aggregation of CLL and NHL is shown, the 6p21.32 association could be reflective of common genetic susceptibility to a range of B-cell LPDs. Despite ~30% of the CLL cases in our study having a family history of CLL or related B-cell LPD we did not, however, find statistically significant support for a disease-locus for CLL within the HLA-DQA1/HLA-DRB5 region.
In conclusion, our analysis provides support for the MHC variation influencing CLL risk and in particular a role of HLA-A*0201 in disease etiology. In terms of impact HLA variation has a weaker effect on CLL risk than the recently identified non-HLA loci(3); this is in stark contrast to HL, a disease which is primarily defined by HLA(15). Finally, although speculative the reciprocal HLA-A*02 associations seen for HL and CLL raise the possibility of differential response to viral infection such as EBV also playing a role in the development of CLL.
Supplementary Material
Supplementary information is available at Leukemia’s website
Acknowledgements
Leukaemia Lymphoma Research provided principal funding for the study. Additional funding was provided by Cancer Research UK and the Arbib fund. We acknowledge National Health Service funding for the Royal Marsden Biomedical Research Centre. Finally we are grateful to all individuals for their participation.
Footnotes
Web Addresses
Human Chromosome 6 Project Overview: http://www.sanger.ac.uk/HGP/Chr6
British 1958 birth cohort: http://www.b58cgene.sgul.ac.uk
NCBI dbSNP: http://www.ncbi.nlm.nih.gov/SNP/
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/
Illumina: http://www.illumina.com/
Golden Helix: http://www.goldenhelix.com/
HLA*IMP: http://oxfordhla.well.ox.ac.uk/hla/
NSCCG:http://www.icr.ac.uk/research/team_leaders/Houlston_Richard/Houlston_Richard_RES/NSCCG/index.shtml
CLL4:http://www.icr.ac.uk/research/research_divisions/Molecular_Pathology/CLL4_trial/index.shtml
Conflict of interest disclosure: The authors declare no competing financial interests.
References
- 1.Sellick GS, Catovsky D, Houlston RS. Familial chronic lymphocytic leukemia. Seminars in oncology. 2006 Apr;33(2):195–201. doi: 10.1053/j.seminoncol.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008 Jan;82(1):48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008 Oct;40(10):1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 4.Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011 Dec 3; doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowther-Swanepoel D, Wild R, Sellick G, Dyer MJ, Mauro FR, Cuthbert RJ, et al. Insight into the pathogenesis of chronic lymphocytic leukemia (CLL) through analysis of IgVH gene usage and mutation status in familial CLL. Blood. 2008 Jun 15;111(12):5691–5693. doi: 10.1182/blood-2008-03-142349. [DOI] [PubMed] [Google Scholar]
- 6.Damle RN, Calissano C, Chiorazzi N. Chronic lymphocytic leukaemia: a disease of activated monoclonal B cells. Best Pract Res Clin Haematol. Mar;23(1):33–45. doi: 10.1016/j.beha.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang SY, Gu HX, Li D, Yang SF, Zhong ZH, Li XK, et al. Association of human leukocyte antigen polymorphism with hepatitis B virus infection and genotypes. Jpn J Infect Dis. 2006 Dec;59(6):353–357. [PubMed] [Google Scholar]
- 8.Souza CF, Noguti EN, Visentainer JE, Cardoso RF, Petzl-Erler ML, Tsuneto LT. HLA and MICA genes in patients with tuberculosis in Brazil. Tissue Antigens. 2011 Oct 28; doi: 10.1111/j.1399-0039.2011.01789.x. [DOI] [PubMed] [Google Scholar]
- 9.Bergamaschi L, Leone MA, Fasano ME, Guerini FR, Ferrante D, Bolognesi E, et al. HLA-class I markers and multiple sclerosis susceptibility in the Italian population. Genes Immun. 2011 Mar;11(2):173–180. doi: 10.1038/gene.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niens M, Jarrett RF, Hepkema B, Nolte IM, Diepstra A, Platteel M, et al. HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV+ Hodgkin lymphoma. Blood. 2007 Nov 1;110(9):3310–3315. doi: 10.1182/blood-2007-05-086934. [DOI] [PubMed] [Google Scholar]
- 11.Urayama KY, Jarrett RF, Hjalgrim H, Diepstra A, Kamatani Y, Chabrier A, et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status-defined subgroups. J Natl Cancer Inst. 2012 Feb 8;104(3):240–253. doi: 10.1093/jnci/djr516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010 Jul;42(7):599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 13.Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009 Aug;41(8):873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010 Aug;42(8):661–664. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat Genet. 2010 Dec;42(12):1126–1130. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.