Fig. 1.
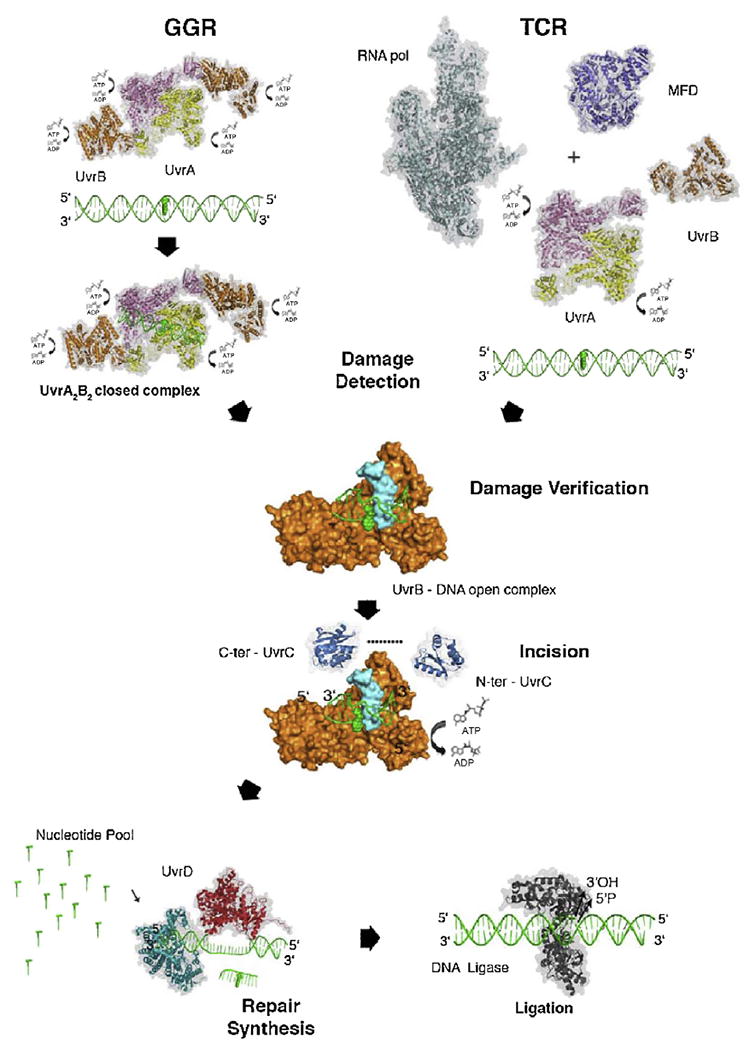
Prokaryotic nucleotide excision repair. Structural model of prokaryotic NER showing the key protein and steps in global genomic repair (GGR) and transcription coupled repair (TCR). TCR damage recognition is initiated by a stalled RNAP (PDB ID: 4LJZ) that recruits MFD (PDB ID: 2EYQ). MFD displaces RNAP and brings UvrA to the damaged site. In GGR, the UvrA2B2 complex (PDB ID: 3UWX) for the contact interface: 3FPN first searches for the distortion along the DNA caused by the lesion. Both pathways converge after the initial recognition steps. UvrA then transfers the damaged DNA to UvrB for damage verification. The dimeric UvrA protein (PDB ID: 2R6F) hydrolyzes both ATP and GTP. It also forms a complex with UvrB (PDB ID: 2FDC) and activates the ATPase activity of UvrB. During damage verification, the β-hairpin of UvrB (shown in turquoise) inserts between the two strands of DNA and forms a stable pre-incision complex, which is believed to activate UvrB’s ATPase. Binding and hydrolysis of ATP by UvrB is essential for recruitment of UvrC. The N-terminal endonuclease domain of UvrC (PDB ID: 1YCZ) initiates the cut 4–5 nucleotides 3′ to the damaged site followed by the 5′ cut by C-terminal endonuclease domain of UvrC (PDB ID: 2NRR) eight nucleotides away from the lesion. UvrD (PDB ID: 2IS1) unwinds the DNA and releases the oligonucleotide containing the lesion. Simultaneously, DNA polymerase I (PDB ID: 2HHQ) synthesizes the missing strand. Finally, DNA ligase I (PDB ID: 1DGS) seals the repair patch. All protein structures in this figure, with the exception of UvrB, are shown with a transparent surface and in ribbon presentation. UvrB is shown with its surface in orange for domains 1 to 3, and the β-hairpin is shown in cyan. C-ter, carboxy terminal; N-ter, amino terminal. From [6] with permission.
