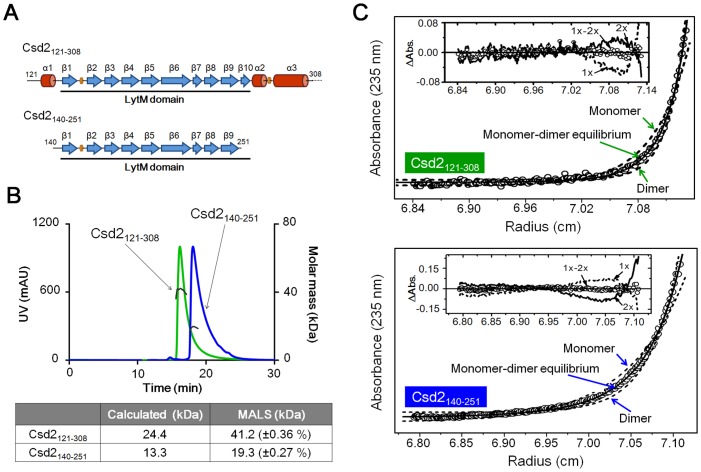
Fig 3. SEC-MALS and equilibrium sedimentation experiments to determine the oligomeric state of H. pylori Csd2 in solution.
(A) Two Csd2 constructs (Csd2121–308 and Csd2140–251) used in these experiments are schematically represented with the secondary structure elements colored as in Fig 1A. Csd2140–251 lacks the helical domain. Csd2121–308 was used for structure determination. (B) SEC-MALS data for two Csd2 protein samples. The black solid lines represent the measured molecular masses. The average molecular masses from MALS analyses are compared with the calculated masses in the table below the chromatography profiles. (C) Equilibrium sedimentation data for two Csd2 protein samples. For Csd2121–308 (top), the circles are experimental data measured at a speed of 35,000 rpm and 5.1 μM protein monomer concentration and the solid line is a fitting line for a reversible monomer-dimer equilibrium model. The two dotted lines are fitting lines for ideal homogeneous monomer and dimer models. Distributions of the residuals for monomer (dotted line), dimer (solid line), and reversible monomer-dimer equilibrium (circles) models are shown in the inset panel. For Csd2140–251 (bottom), the circles are experimental data measured at a speed of 35,000 rpm and 14.5 μM protein monomer concentration and the solid line is a fitting line for a reversible monomer-dimer equilibrium model. The two dotted lines are fitting lines for ideal homogeneous monomer and dimer models. Distributions of the residuals for monomer (dotted line), dimer (solid line), and reversible monomer-dimer equilibrium (circles) models are shown in the inset panel. Equilibrium sedimentation data indicate that both Csd2121–308 and Csd2140–251 are in reversible monomer-dimer equilibrium in solution.