Fig 7. The C-terminal tail of Csd2 (chain B’) is bound to the substrate-binding groove in the LytM domain of Csd1 (chain C) in Csd1-Csd2 dimer I.
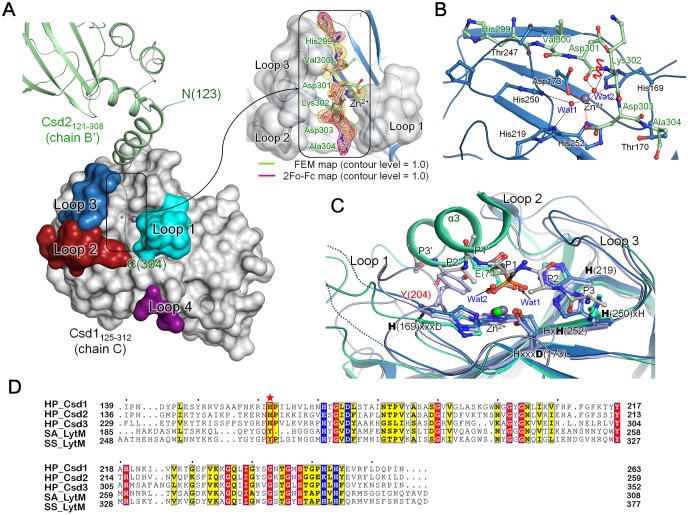
(A) In the structure of Csd1-Csd2 dimer I, the C-terminal residues (His299−Ala304) of Csd2121–308 from an adjacent asymmetric unit (chain B’ shown in ribbon diagram) occupy the substrate-binding groove of the LytM domain in Csd1125–312 (chain C shown in surface diagram). Four loops of Csd1 LytM domain that form the substrate-binding groove are labeled and colored as in Fig 5. The ribbon diagram is colored as in S1 Fig A close-up view on the right represents the Csd2 C-terminal tail residues located in the substrate-binding groove of Csd1 LytM domain. The Csd2 tail residues (enclosed in the black box) are shown in a stick model, with the electron density shown in mesh. The electron density for the Csd2 tail in the feature-enhance map (FEM) calculated by using PHENIX program [47] (lime colored mesh) and 2mFo − DFc map (magenta colored mesh) are shown at the 1.0 σ level. (B) A detailed view of the interactions between the Csd2 tail residues and the substrate-binding groove of the Csd1 LytM domain (shown in ribbon diagram, colored as in S1 Fig). Both main chains and side chains of the Csd2 tail residues are shown in a stick model, with the candidate peptide bond that might be cleaved by the enzymatic activity of Csd1 is indicated by a red wavy line. Side chains of Csd1125–312 residues interacting with the Csd2 tail residues are shown in a stick model. Grey and red spheres represent the Zn2+ ion and water molecules, respectively. Zn2+-coordination (canonical) and hydrogen bonds with waters are indicated by red and black dotted lines, respectively. (C) Superposition of LytM domains in H. pylori Csd1 (skyblue), H. pylori Csd3 (light green; PDB code, 4RNY), and S. aureus LytM bound with tetraglycine phosphinate (purple; PDB code, 4ZYB) shows that the two water molecules (Wat1 and Wat2) of Csd1125–312 chain C of heterodimer I overlap nicely with side chain oxygen atoms of Glu74 (labeled in light green) from the helix α3 of the inhibitory Domain 1 in Csd3 and also with those of the phosphinate molecule (black). The Csd2 tail is simplified as a poly-alanine model (grey). The bound Zn2+ ions are indicated by grey, purple, and green spheres for H. pylori Csd1, H. pylori Csd3, and S. aureus LytM, respectively. Two dotted lines represent the disordered regions in Loop 1 of Csd1. The metal-coordinating residues in the H(169)xxxD(173) and HxH(252) motifs and the conserved catalytic residues in the H(250)xH motif and an additional catalytic histidine residue H(219) of Csd1, as well as corresponding residues of H. pylori Csd3 and S. aureus LytM, are shown in a stick model. Tyr204 (labeled in red) of S. aureus LytM is shown in a stick model. (D) Sequence alignment of LytM domains in Csd1, Csd2, and Csd3 from H. pylori 26695 strain [Csd1 (HP_Csd1; SWISS-PROT accession code O26068), Csd2 (HP_Csd2; O26069), and Csd3 (HP_Csd3; O25247)], S. aureus LytM (SA_LytM; O33599), and S. simulans lysostaphin (SS_LytM; P10547). Tyr204 of S. aureus LytM is marked by a red star. Conserved residues of the characteristic motifs are colored in blue.