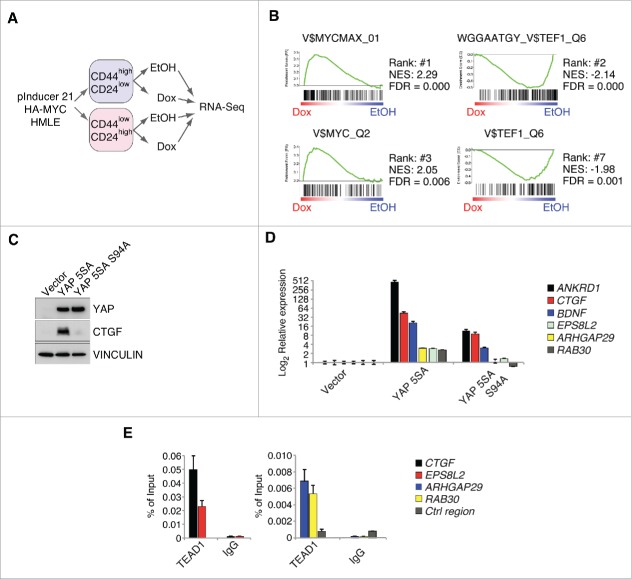
Figure 1.
MYC restrains TEAD-dependent YAP functions. A: Workflow for the RNA-Sequencing experiment performed in FACS-sorted HMLE cells expressing a doxycycline-inducible HA-MYC allele (pInducer21-HA-MYC). B: Gene set enrichment analysis for the C3 gene set (motifs) in the CD44high/CD24low HMLE population 8 hours after MYC induction. MYC and MYCMAX represent potential binding sites for the MYC/MAX heterodimer, whereas TEF1 represents predicted binding sites for TEAD1 (also called TEF1). NES = normalized enrichment score; FDR = false discovery rate. C: CD44high/CD24low HMLE cells were infected with the indicated YAP 5SA mutants (YAP 5SA or the TEAD-binding compromised YAP 5SA S94A mutant) and analyzed by immunoblots. VINCULIN served as a loading control. D: qRT-PCR analyses of the direct YAP/TAZ target genes ANKRD1 and CTGF, as well as additional genes (BDNF, ARHGAP29 and RAB30) from the WGGAATGY_V$TEF1_Q6 C3 gene set. E: qChIP analysis in MCF7 cells for TEAD binding to the promoters of the given genes. IgG served as a negative control. Ctrl region = U2 promoter region.