Summary
G protein-coupled receptors (GPCRs) mediate cellular responses to a wide variety of extracellular stimuli. GPCR dimerization may expand signaling diversity and tune functionality, but little is known about the mechanisms of subunit assembly and interaction or the signaling properties of heteromers. Using single molecule subunit counting on Class C metabotropic glutamate receptors (mGluRs), we map dimerization determinants and define a heterodimerization profile. Intersubunit fluorescence resonance energy transfer measurements reveal that interactions between ligand binding domains control the conformational rearrangements underlying receptor activation. Selective liganding with photoswitchable tethered agonists conjugated to one or both subunits of covalently linked mGluR2 homodimers reveals that receptor activation is highly cooperative. Strikingly, this cooperativity is asymmetric in mGluR2/mGluR3 heterodimers. Our results lead to a model of cooperative activation of mGluRs that provides a framework for understanding how class C GPCRs couple extracellular binding to dimer reorganization and G protein activation.
Introduction
G protein coupled receptors (GPCRs) respond to a wide array of extracellular stimuli to initiate intracellular signaling via G proteins and arrestins (Pierce et al., 2002). Assembly into homo- or hetero-oligomeric complexes has emerged as a potentially crucial aspect of GPCR function, which can modulate sensitivity to stimuli, basal activity, effector coupling, ligand bias, kinetics, and subcellular targeting (Gurevich and Gurevich, 2008; Lohse, 2010) (Ferre et al., 2014). Breakthrough studies have provided snapshots of GPCR structures in distinct conformations (Rasmussen et al., 2011; Katritch et al., 2013; Venkatakrishnan et al., 2013) and revealed that they are extremely dynamic (Nygaard et al., 2013; Manglik et al., 2015; Sounier et al., 2015; Isogai et al., 2016). This has led to the emerging idea that conformational dynamics are central to ligand recognition, activation and signaling by GCPRs. However, little is known about the impact of oligomerization on GPCR conformational dynamics and whether or how this confers cooperativity onto signaling.
Despite the interest in GPCR oligomerization, complexes have been difficult to observe and analyze structurally or biophysically within the class A and B families of GPCRs (Vischer et al., 2015). However, class C GPCRs, which are characterized by a large extracellular ligand binding domain (LBD), have been shown to assemble into stable dimers or higher order oligomers both in crystal structures of isolated LBDs (Geng et al., 2013; Kunishima et al., 2000) and biochemical or spectroscopic analyses of full length proteins (Romano et al., 1996; Doumazane et al., 2011; Maurel et al., 2008; Comps-Agrar et al., 2011).
Metabotropic glutamate receptors (mGluRs) are class C GPCRs that modulate synaptic strength and serve as drug targets for neurological disorders (Niswender and Conn, 2010). mGluRs consist of three domains: an N-terminal bi-lobed LBD, a 7 helix trans-membrane domain (TMD) and a cysteine rich domain (CRD) that links the LBD to the TMD. Dimerization of full length mGluRs is required for G protein activation (El Moustaine et al., 2012) and has been shown to be partially mediated by an intersubunit disulfide bridge between the LBDs (Romano et al., 1996). Biochemical and spectroscopic studies have shown that activation coincides with intersubunit reorientation of both the extracellular and transmembrane domains of the receptor (Doumazane et al., 2013; Olofsson et al., 2014; Vafabakhsh et al., 2015; Kunishima et al., 2000; Tateyama et al., 2004; Xue et al., 2015). However, the mechanisms of dimerization and the role of dimer interfaces in activation, intersubunit communication and mGluR function are not clear.
In this study we use quantitative subunit counting to find that mGluR homo- and hetero-dimerization depends primarily on interactions at a hydrophobic interface in the upper lobe of the LBD, with modest contributions from an intersubunit disulfide bridge and TMD interaction. Fluorescence resonance energy transfer (FRET) enables us to quantify the contribution of interface hot spots to global conformational dynamics. An approach to targeted, single subunit activation, using photoswitchable tethered ligands in dimers of defined stoichiometry, enables us to unravel the mechanism of cooperativity in mGluR activation. We find that cooperativity in dimers gives rise to boosted activity and that heterodimerization gives rise to the emergence of unique biochemical properties. Our observations lead to a model of mGluR gating that accounts for cooperativity in basal activity in the absence of agonist as well as partial and full activation across the concentration range of agonist in both homo- and heterodimers.
Results
Dimerization is primarily mediated by interactions between ligand binding domains
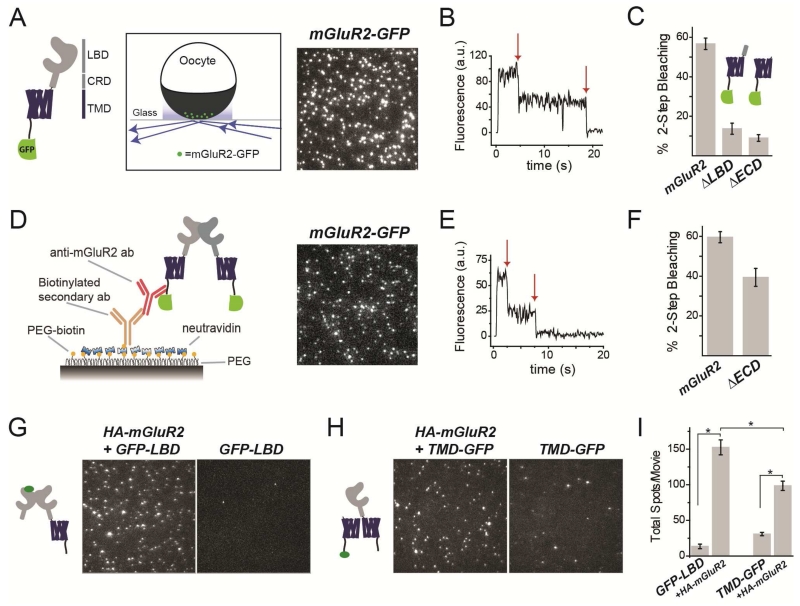
Previous studies of mGluR oligomerization employed classical biochemical methods to assay denatured proteins in vitro (Ray and Hauschild, 2000; Romano et al., 1996; Tsuji et al., 2000); (Romano et al., 2001) or ensemble (macroscopic) time-resolved FRET on receptors in the plasma membrane of live cells, which relies on theoretical models and is sensitive to conformational changes (Doumazane et al., 2011). To analyze mGluR stoichiometry, we used single molecule subunit imaging of individual protein complexes to count photobleaching steps in green fluorescent protein (GFP)-tagged receptors. We expressed a C-terminally GFP-tagged mGluR2 (mGluR2-GFP) in Xenopus oocytes at low density to ensure that individual complexes would be spatially resolved as individual fluorescent spots (Fig. 1A; Fig. S1A). Using total internal reflection (TIRF) microscopy to confine excitation to the plasma membrane, we observed GFP photobleaching (Fig. 1B) and found that all spots bleached in one or two steps, with ~60% bleaching in two steps (Fig. 1C, Fig. S1A). This is consistent with mGluR2 being a dimer, with a probability of ~0.7 that its GFP is fluorescent (Fig. S1A), due to 20-30% of GFPs which either mis-fold or fail to undergo chromophore maturation (Nakajo et al., 2010; Ulbrich and Isacoff, 2007). Importantly, mGluR2-GFP proved to be functional, supporting glutamate-induced currents from co-expressed G protein activated inward rectifier potassium (GIRK) channels (Fig. S1A). Similar photobleaching distributions were obtained with GFP-tagged versions of mGluR3 (group II), mGluR7(group III), and mGluR1 and mGluR5 (group I); and each of these was also functional, producing glutamate-induced GIRK current (mGluR3 and 7) or calcium-activated chloride current (mGluR1 and 5) (Fig. S1B-E).
Figure 1. Dimerization of mGluR2 is mediated primarily by the ligand binding domain.
(A) Left, domain structure of the mGluR2-GFP construct. LBD=Ligand Binding Domain; CRD=Cysteine Rich Domain; TMD=Transmembrane Domain. Right, schematic showing TIRF image of single mGluR2-GFP molecules in the plasma membrane of oocytes.
(B) Representative photobleaching trace for a single mGluR2-GFP complex. Arrows show photobleaching steps.
(C) Summary of photobleaching step analysis for mGluR2-GFP and truncations. ~60% 2-step photobleaching is consistent with a strict dimer with ~75% GFP maturation.
(D) Left, Schematic showing SimPull technique for pulldown with an anti-mGluR2 antibody. Right, representative TIRF image of single mGluR2-GFP molecules isolated from HEK293T cell lysate.
(E) Representative photobleaching trace for a single mGluR2-GFP complex.
(F) Summary of photobleaching step analysis for mGluR2-GFP and ΔECD-GFP in SimPull.
(G-H) TIRF images of single GFP-LBD (G) or TMD-GFP (H) subunits isolated using an anti-HA antibody in the presence or absence of full length HA-mGluR2.
(I) Summary of pull down efficiency for GFP-LBD and TMD-GFP in the absence or presence of HA-mGluR2. (Unpaired t-test, p=0.00003 between GFP-LBD with and without HA-SNAP; p=0.0001 between TMD-GFP with and without HA-SNAP; p=0.004 between GFP-LBD and TMD-GFP.)
Error bars show S.E.M. calculated from multiple experiments (N>3).
We next asked which domains mediate dimerization. We truncated mGluR2-GFP by removing only the LBD (ΔLBD-GFP) or the entire extracellular domain (ΔECD-GFP). In both cases we observed a near-complete reduction in 2-step photobleaching (Fig 1C; Fig. S1F, G), indicating that the extracellular domain, particularly the LBD, is required for efficient dimerization.
Subunit counting in Xenopus oocytes requires low expression levels in order to resolve single molecules in the plasma membrane. To analyze stoichiometry under high density expression, we expressed mGluR2-GFP under the CMV promoter in HEK293T cells (Fig. S1H) and isolated receptors in detergent using the SimPull technique (Jain et al., 2011) to obtain a low density of immune-purified protein on a passivated surface (Fig. 1D) (Vafabakhsh et al., 2015). Consistent with results from oocytes, ~60% of mGluR2-GFP spots bleached in two steps in SimPull (Fig. S1H). ΔECD-GFP expressed at lower levels and had poorer surface localization compared to mGluR2-GFP (Fig. S1I). Interestingly, although deletion of the extracellular domain reduced 2-step photobleaching (Figs. 1F, S1I), this reduction was less extreme than what was seen in Xenopus oocytes (Fig. 1C). ΔECD-GFP constructs for both mGluR3 and mGluR1 showed a similar behavior (Fig. S1J). These observations suggest that, at the high expression density obtained in HEK293T cells, parts of the protein other than the ECD contribute to dimerization. This interpretation was supported by the finding that SimPull from oocytes injected with 100-fold higher levels of RNA for ΔECD-GFP leads to an increase in the percentage of 2-step photobleaching spots (~30% vs. <5%) (Fig. S1K).
To further probe the roles of the LBD and the TMD in dimerization, we examined the ability of truncated domains to co-assemble with full length mGluR2. Co-expression of either “GFP-LBD” or “TMD-GFP” (ΔECD-mGluR2-GFP) with full length HA-mGluR2, followed by SimPull with anti-HA antibodies led to immobilization of GFP spots (Fig. 1G,H) with more efficient co-assembly of mGluR2 with the isolated LBD than with the isolated TMD (Fig. 1I). Most spots bleached in a single step confirming that one full length mGluR2 subunit co-assembles with a single isolated domain (92.4 ± 0.1% for GFP-LBD and 89.6 ± 0.8% for TMD-GFP). These experiments confirm that the primary dimer interface is the LBD, with the TMD representing a secondary interface.
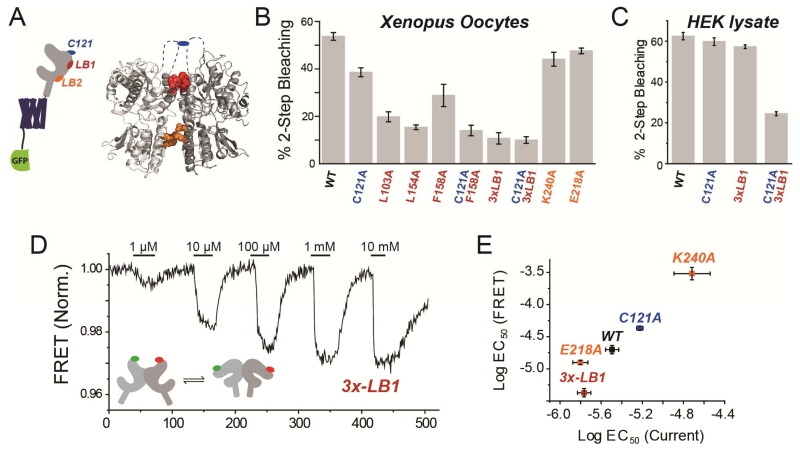
Given the primary role of the LBD in dimerization, we investigated the inter-LBD dimer interface by examining crystal structures of the isolated LBDs of mGluRs (Fig. 2A) (Kunishima et al., 2000; Muto et al., 2007; Tsuchiya et al., 2002). In both the relaxed and active states, a cluster of conserved hydrophobic residues in helices B and C lie at an interface between the upper LBD lobes (LB1) (Fig. 2A; Fig. S2A, B). In the active state, an additional interface is observed between conserved charged residues in the lower LBD lobes (LB2), where Gd3+ has been shown to bind in mGluR1 (Tsuchiya et al., 2002) (Fig. 2A; Fig. S2C). Furthermore, a conserved cysteine (C121 in mGluR2), located in the structurally unresolved loop between helices B and C, has been shown to be crucial for dimerization in denaturing gels (Ray and Hauschild, 2000; Romano et al., 1996; Tsuji et al., 2000); (Romano et al., 2001) (Fig. 2A; Fig. S2A). We mutated residues at these potential interfaces and found a major reduction in dimerization in the plasma membrane of Xenopus oocytes with mutations at the LB1 interface, but only a minor reduction with a mutation of C121 to alanine (Fig. 2B; Fig. S2D; Unpaired T-tests, p=0.003 between C121A and L103A, p=0.0003 between C121A and L154A, p=0.00006 between C121A and 3xLB1). Consistent with the weak effect of C121, application of DTT, to reduce a potential inter-subunit disulfide bond, did not alter receptor stoichiometry in wild type (WT) mGluR2-GFP (Fig. S2E). However, in the background of the single mutation F158A to the hydrophobic LB1 interface, the additional mutation of C121A further reduced dimerization (Fig. 2B). In contrast, LB2 mutants E218A and K240A did not significantly alter dimerization (Fig. 2B). Under high-density expression in HEK293T cells, followed by SimPull purification, dimerization was only reduced when all three LB1 mutations (“3xLB1”) were combined with the C121A mutation (Fig. 2C). We also analyzed expression and surface targeting of dimer interface mutants using confocal imaging. C-terminally GFP-tagged versions of mGluR2wt, 3xLB1, C121A, and 3xLB1/C121A showed clear membrane enrichment and similar levels of fluorescence (Fig. S2F). We quantified surface targeting with membrane impermeable dye conjugation to N-terminally SNAP-tagged variants and found similar levels for all variants tested (Fig. S2G,H). Together these experiments show that dimerization is mediated primarily by the hydrophobic LB1 interface, along with a weak contribution from a covalent intersubunit disulfide bridge, and little contribution from the LB2 interface.
Figure 2. Mutational analysis of mGluR2 dimer interface: assembly and functional effects.
(A) Schematic, left, and crystal structure of mGluR1 in the “active” state (PDB: 1EWK), right, showing 3 regions proposed to form the LBD dimer interface.
(B-C) Summary of stoichiometry of dimer interface mutants in oocytes (H) and SimPull from HEK293T cell lysate (I). “3x-LB1” is the construct containing L103A, L154A, and F158A mutations.
(D) Representative FRET trace showing glutamate-induced reductions in intersubunit FRET between LBDs for N-terminally SNAP and CLIP-tagged versions of mGluR2-3xLB1.
(E) Summary of glutamate EC50 determinations from activation of GIRK channels (current) versus LBD conformational change (FRET) in mGluR2WT (WT) and dimer interface mutants.
Error bars show S.E.M. calculated from multiple experiments (N>3).
Role of mGluR2 homodimer LBD interface in conformational dynamics and activation
We next asked how the LBD dimer interfaces affect receptor function by recording glutamate-evoked currents from HEK293T cells co-expressing WT or mutant versions of mGluR2 along with the GIRK channel. Interestingly, we found that the mGluR2-C121A mutation reduced apparent glutamate affinity (Fig. S2I), whereas weakening of the LB1 interface with the triple mutant 3xLB1 increased apparent glutamate affinity (Fig. S2I). To directly measure the activation-associated conformational changes of the LBDs, we performed an ensemble intersubunit FRET assay in HEK293T cells using mGluRs that were fused at their N-terminus to SNAP- or CLIP-tags and conjugated to acceptor (Alexa-647) or donor (DY-547) fluorophores (Doumazane et al., 2013), respectively. As shown earlier, glutamate binding induces closure and reorientation of the LBD that increase distance between the fluorophores, thus reducing FRET (Doumazane et al., 2013). Consistent with the GIRK assay, glutamate dose-response curves of the conformational changes associated with activation revealed that the C121A mutant reduces apparent affinity and the 3xLB1 triple mutant increases apparent affinity (Fig. 2D, E; S2J). Neutralization of charged residues on helix F in the LB2 interface also had an effect: K240A strongly decreased the apparent glutamate affinity, as recently reported (Vafabakhsh et al., 2015), while E213A, E218A, and E222A weakly increased apparent affinity (Fig. 2E; S2K, L) (Unpaired T-test with mGluR2; p=0.0034 for E213A, p=0.0037 for E218A, and p=0.28 for E222A).
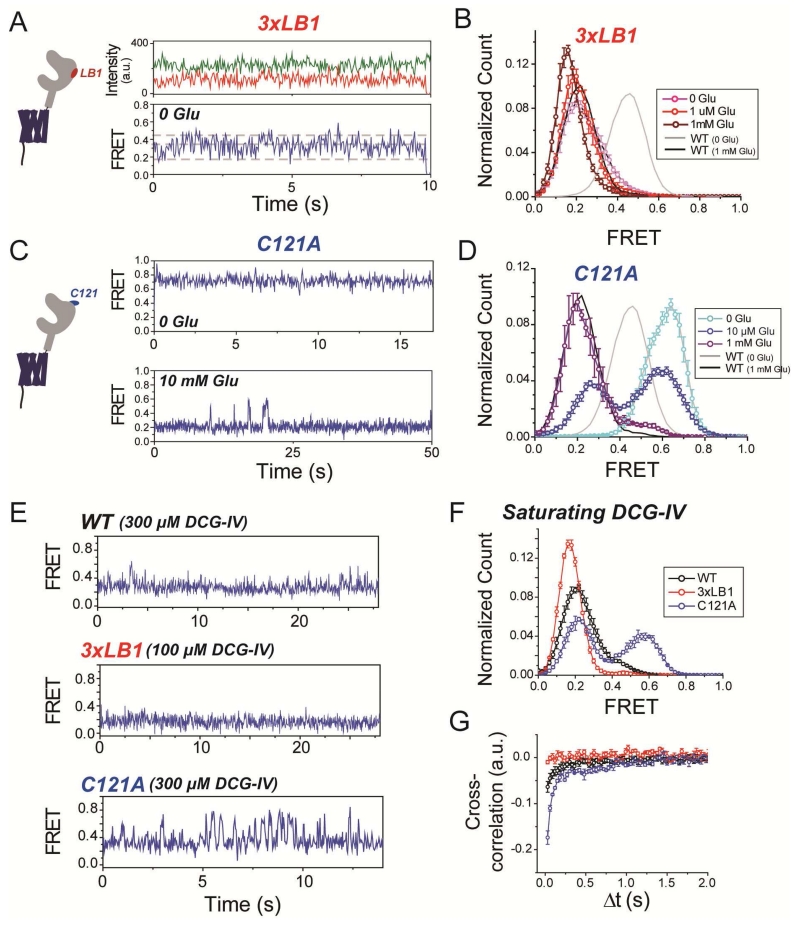
To better understand the role of the dimer interfaces in activation, we turned to single molecule FRET (smFRET), which recently revealed that group II mGluRs transition between three states with distinct FRET efficiencies: ~0.45 (“high”), ~0.35 (“medium”), and ~0.2 (“low”), with the low FRET state corresponding to the active conformation (Vafabakhsh et al., 2015). As seen before, in the absence of glutamate mGluR2WT shows few transitions out of the high FRET state, and in saturating glutamate few transitions out of the low FRET state, but at intermediate concentrations it shows rapid dynamics (Fig. S3A-C). In contrast, in the absence of glutamate, mGluR2-3xLB1 showed transitions from the inactive high FRET state to the intermediate and low FRET states (Fig. 3A), with predominant occupancy of the activated state (Fig. 3B). Glutamate application further increased occupancy of the low FRET state (Fig. S3D), fully occupying it at saturating concentrations (Fig. 3B) and, similar to mGluR2WT, abolishing dynamics, as measured by donor and acceptor cross-correlation (Fig. S3E). Interestingly, this lowest FRET state of mGluR2(3xLB1) was lower in FRET than what was observed in mGluR2WT (Fig. 3B, S3F), suggesting that 3xLB1 stabilizes a state with increased degrees of LBD closure and/or dimer reorientation. These observations suggest that the hydrophobic LB1 interface provides energy to prevent spontaneous LBD closure and dimer reorientation in the absence of agonist.
Figure 3. smFRET analysis of mGluR2 LBD interface mutants.
(A) smFRET traces of mGluR2-3xLB1 showing spontaneous dynamics in the absence of glutamate. Top, donor (green) and acceptor (red) fluorescence for a single mGluR2-3xLB1 dimer. Bottom, smFRET calculated from donor and acceptor fluorescence values in top traces. Dotted gray lines show the 3 FRET states obtained from Gaussian fits of smFRET histograms.
(B) Histogram showing smFRET distributions for mGluR2-3xLB1. Distributions for mGluR2WT in 0 (grey) or 1 mM glutamate (black) are shown as solid lines for comparison.
(C) Representative smFRET traces of mGluR2-C121A, in the absence of glutamate (top) and for mGluR2-C121A showing transitions to resting conformation in the presence of saturating 10 mM glutamate (bottom).
(D) Histogram showing smFRET distributions for mGluR2-C121A. Note the small increase in high FRET population for C121A in the presence of saturating glutamate compared to WT.
(E) Representative smFRET trace for WT, 3xLB1, and C121A showing the level of dynamics in the presence of saturating DCG-IV (100 uM for C121A; 300 uM for WT, 3xLB1).
(F) Histogram showing smFRET distributions for WT, 3xLB1, and C121A in the presence of saturating DCG-IV.
(G) Cross-correlation plots showing relative dynamics for WT, 3xLB1, and C121A in the presence of saturating DCG-IV.
Error bars show S.E.M. calculated from multiple experiments (N≥3).
In contrast, in the absence of glutamate, mGluR2(C121A) displayed minimal dynamics and was shifted to a higher peak FRET of the inactive conformation (~0.6 vs. ~0.45; Fig. 3C, D). Application of glutamate increased the population of the low FRET conformation, but with a lower apparent affinity than seen in WT mGluR2 (Fig. 3D; Fig. S3G, H). Even in saturating glutamate (10 mM) mGluR2(C121A) showed dynamics out of the low FRET state (Fig. 3C, bottom; Fig. S3I). These observations suggest that the intersubunit disulfide bridge primes the resting conformation for the activation rearrangement and stabilizes the active state.
To further examine the differences between the hydrophobic LB1 (“3xLB1”) and covalent (“C121A”) interfaces, we performed smFRET experiments with the partial agonist DCG-IV which we previously showed to have partial efficacy due to partial active state occupancy at saturating concentrations (Vafabakhsh et al, 2015). Compared to mGluR2WT, at saturating DCG-IV, mGluR2(C121A) showed a much reduced occupancy of the low FRET state, while mGluR2(3xLB1) showed a greater low FRET state occupancy (Fig. 3E, F). Consistent with this, dynamics at saturating DCG-IV were abolished in mGluR2(3xLB1) and increased in mGluR2(C121A) (Fig. 3E, G). These results support the interpretation that the active state is stabilized by the 3xLB1 mutation and destabilized by the C121A mutation.
We also measured smFRET from the mGluR2(3xLB1/C121A) quadruple mutant. Similar to mGluR2(3xLB1), the quadruple mutant showed spontaneous dynamics, including visits to the active state (Fig. S3J, K), indicating that an intact disulfide bond is not required for the introduction of basal conformational dynamics by the 3xLB1 mutant. However, like mGluR2(C121A), the quadruple mutant showed an increase in FRET values associated with inactive states, as well as incomplete population of the active state in saturating glutamate (Fig. S3J, K). In saturating DCG-IV, mGluR2(3xLB1/C121A) populated the low FRET state at a level intermediate between that of the 3xLB1 or C121A mutant alone (Fig. S3M), and had a slight decrease in low FRET peak value, similar to 3xLB1 (Fig. S3M). Together these observations indicate that mutations to LB1 and the disulfide interface have effects that do not occlude one another, suggesting that they act through distinct mechanisms.
Having established the roles of dimer interfaces in the assembly and activation of mGluR2, we next turned to an analysis of heteromeric combinations of mGluR2.
Heterodimerization of mGluR2: preference for mGluR3 and role of the LBD
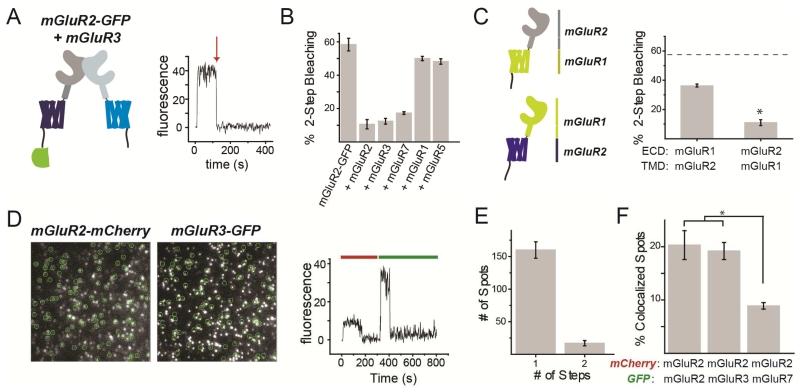
Initial biochemical studies did not find evidence for mGluR heteromerization between subtypes (Romano et al., 1996), but recent spectroscopic and biochemical studies have indicated that mGluRs can heteromerize, although there has not been complete agreement about which combinations are possible (Delille et al., 2012; Doumazane et al., 2011; Yin et al., 2014). To assess the ability of mGluR2 to heteromerize with other subtypes, we turned again to single molecule subunit counting. We first co-expressed mGluR2-GFP with excess untagged mGluRs and measured the ability of the untagged mGluR to assemble with mGluR2-GFP to create heterodimers with only one GFP. For example, co-expression of mGluR2-GFP with untagged mGluR3 (Fig. 4A) reduced 2-step photobleaching as drastically as did co-expression with untagged mGluR2 (Fig. 4B). Untagged mGluR7 was nearly as potent, but group I members mGluR1 and 5 had almost no effect (Fig. 4B). These observations agree with recent findings that indicated that group II/III mGluRs cannot co-assemble with group I mGluRs (Doumazane et al., 2011). Consistent with this, in SimPull, mGluR2 pulled down mGluR3-GFP but not mGluR1-GFP (Fig. S4A, B). Moreover, the mGluR3-GFP spots pulled down by mGluR2 showed single-step photobleaching (Fig. S4C), confirming the strict heterodimerization.
Figure 4. mGluR2 heterodimerizes with Group II/III mGluRs and prefers intra- over inter-group assembly.
(A-B) Coexpression of excess untagged group II or III mGluRs decreases 2-step photobleaching of mGluR2-GFP in Xenopus oocytes.
(C) Co-expression of chimeras between mGluR1 and mGluR2 (left) decreases 2-step photobleaching with a stronger effect when the ECD is from mGluR2 rather than mGluR1 (right). * indicates statistical significance (unpaired t-test, p=.0002). Dotted line shows the level of 2-step bleaching observed for mGluR2-GFP alone.
(D) Images (left) showing colocalization between mGluR2-mCherry and mGluR3-GFP and representative trace (right) showing 1-step photobleaching in red and green.
(E) Photobleaching step analysis showing primarily 1-step GFP photobleaching for mGluR3-GFP in complex with mGluR2-mCherry.
(F) Summary of colocalization analysis for mGluR2-mCherry with either mGluR2-GFP, mGluR3-GFP, or mGluR7-GFP. * indicates statistical significance (unpaired t-test, p=0.0003 between mGluR2 and mGluR7 and p=0.0007).
Error bars show S.E.M. calculated from multiple experiments (N≥3).
To identify the domains that mediate heterodimer specificity, we made chimeras between mGluR2 and mGluR1 which contain the ECD of one and the TMD (and C-terminal) of the other (Fig. 4C). In oocytes and SimPull, both chimeras remained strict dimers (Fig. S4D, E). When co-expressed in excess with mGluR2-GFP in Xenopus oocytes, both chimeras decreased the 2-step photobleaching of mGluR2-GFP, but this effect was significantly stronger for the construct containing the mGluR2 ECD (Fig. 4C). Consistent with this, in SimPull, mGluR2 was able to pull down both chimeras, but was significantly more efficient at pulling down the chimera with the mGluR2 ECD (Fig. S4F-H).
To directly visualize heterodimers and assess their relative affinities, we performed 2-color photobleaching experiments in Xenopus oocytes. We co-expressed mGluR2-mCherry with either mGluR2-GFP, mGluR3-GFP, or mGluR7-GFP at similar surface densities, identified co-localized red and green spots (Fig. 4D) and counted photobleaching in the green channel (Fig. 4E). mGluR2-mCherry co-localized with all 3 variants to form strict heterodimers (Fig. 4E) but a higher percentage of co-localized spots were found with mGluR2-GFP or mGluR3-GFP compared to mGluR7-GFP (Fig. 4F). Together, these experiments show that mGluR2 can heterodimerize with equal affinity with the other group II mGluR, mGluR3, as with itself, as well as with a group III mGluR, albeit with a lower preference.
The importance of the dimer interface in receptor activation (Fig. 2, 3), suggests that subunit interaction and cooperativity may operate in mGluR2. Furthermore, while smFRET analysis has revealed three intersubunit conformations, it remains unclear how occupancy by zero, one or two ligands in a dimer drives occupancy of those states.
Photoswitchable tethered ligands and linked dimers reveal cooperativity of mGluR2
Having seen that mGluRs dimerize via inter-subunit LBD interactions, and that these interactions are important for activation, we asked whether LBD-LBD interaction introduces cooperativity into the activation process. Initial work with radioligand binding and native tryptophan fluorescence suggested negative cooperativity of glutamate binding in isolated mGluR1 LBDs (Suzuki et al., 2004). Ensuing work suggested that binding of glutamate to one subunit can activate mGluRs, but that binding to both subunits activates more efficiently (Kniazeff et al., 2004), although another study suggested that activation does not occur until 2 ligands bind (Kammermeier and Yun, 2005). These studies faced the difficulty of confining ligand binding to one subunit by using mutations to lower agonist affinity in the other subunit. However, it was not possible to exclude the possibilities that low affinity mutants permit short-lived binding, that the affinity or gating of the mutant subunit is influenced by co-assembly with a wildtype subunit, or that the mutation could itself alter conformational energetics. For these reasons, we turned to the photoswitchable tethered ligand (PTL) approach in which, a cysteine residue on the outer, solvent facing surface of the LBD covalently anchors a “ligand on a string,” which can be rapidly and reversibly photoisomerized to place the ligand in the orthosteric binding pocket and thereby agonize or antagonize only that subunit. This approach has been used in a variety of neurotransmitter receptors (Volgraf et al., 2006; Reiner et al., 2015; Lin et al., 2015), including mGluRs (Levitz et al., 2013; Carroll et al., 2015).
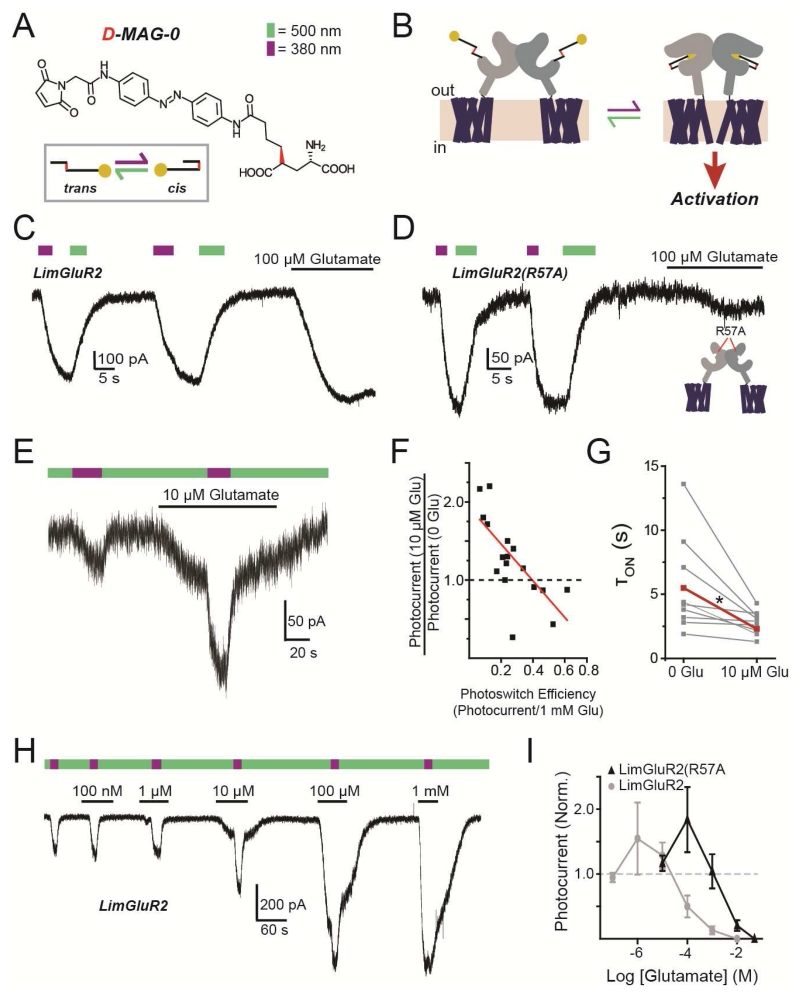
The glutamate-presenting PTL D-MAG-0 (“MAG”) (Fig. 5A) can be covalently attached to a cysteine substituted onto the solvent exposed surface of the LBD of mGluR2 (mGluR2(L300C)) to form “LimGluR2” (Fig. 5B) (Levitz et al., 2013), which is activated rapidly, and with high efficacy by light, as seen by the induction of GIRK current in HEK293 cells (Fig. 5C). A similar MAG PTL has been shown in an ionotropic glutamate receptor to function as a high occupancy agonist with a high local concentration (Gorostiza et al., 2007). To test if this is the case with mGluR2, we lowered the glutamate affinity of LimGluR2 with the mutation R57A (Malherbe et al., 2001) (Fig. S5A) and still observed large photocurrents with similar efficacy compared to saturating glutamate (Fig. 5D; Fig. S5B). LimGluR2(R57A) also showed a reduced affinity for the antagonist LY341495, but retained block by a negative allosteric modulator (NAM) which binds in the TMD (Fig. S5E-J). The efficient photoactivation of LimGluR2(R57A) confirms that MAG operates as a high efficacy, high occupancy agonist that makes it well-suited to study receptor cooperativity.
Figure 5. Photoactivation of tethered ligands reveals cooperative activation in mGluR2.
(A) Chemical structure of the D-MAG-0 photoswitchable tethered ligand. Irradiation with 500 nm light (green arrow) induces the trans-configuration and 380 nm light (violet arrow) induces the cis-configuration.
(B) Schematic of photoactivation of mGluR2 that is conjugated to D-MAG-0 (“LimGluR2”).
(C) Representative HEK293T whole-cell recording where LimGluR2 is co-expressed with GIRK1(F137S) as a reporter. 380 nm light (violet bar) induces an inward current that is turned off by 500 nm light (green bar), compared to current evoked by 100uM glutamate.
(D) Low affinity LimGluR2(R57A) shows large photocurrents and diminished glutamate response.
(E) Partial D-MAG-0 labeling yields weak LimGluR2 photocurrent that is potentiated by a low concentration of glutamate.
(F) Summary of photocurrent potentiation (y-axis) by 10 μM glutamate as a function of degree of D-MAG-0 labeling (photoswitch efficiency; x-axis). Red line shows linear fit.
(G) Summary of accelerating effect of 10 μM glutamate on photocurrent kinetics. Individual cells (gray) in two conditions connected by lines, with average (red). * indicates statistical significance (paired t-test, p=0.007).
(H-I) Concentration-dependence of glutamate-mediated photocurrent potentiation for LimGluR2 and LimGluR2(R57A), showing representative trace from cell expressing LimGluR2 (H) and average relation (I).
Error bars show S.E.M. calculated from multiple experiments (N≥3).
First, in order to generate a distribution of dimers in each cell, with either zero, one or two attached MAGs, we under-labeled LimGluR2. Under these conditions, glutamate at or below the dissociation constant is expected to bind a fraction of the subunits, whether their LBDs are attached to MAG or not. In contrast, light will only induce liganding in MAG-labeled subunits and dimers with no labeled subunits will not respond. We found that photoswitching to the activating cis state evoked a small photocurrent, consistent with sparse MAG conjugation, but that the photocurrent was usually potentiated by prior application of a sub-saturating concentration of glutamate (Fig. 5E). The magnitude of photocurrent potentiation by glutamate depended on the extent of MAG labeling, which we determined from a measure of photoswitch efficiency, with lower MAG labeling giving greater potentiation (Fig. 5F). Photocurrent amplitude potentiation was accompanied by an increase in the speed of photocurrent activation (Fig. 5G). Since photo-agonism by MAG is equipotent to activation by glutamate (Levitz et al., 2013), MAG-conjugated LBDs that bind glutamate should maintain the same occupancy when MAG is photoswitched into the binding site, displacing free glutamate, with no effect on GIRK activation. At the same time, dimers with neither LBD labeled with MAG will not respond to light. Thus, the potentiating effect of sub-saturating glutamate on the photocurrent must be due to receptors in which only one subunit is labeled with MAG, and must occur when glutamate binds to the unlabeled subunit while MAG photo-agonizes the labeled subunit. Consistent with this, photocurrent potentiation in LimGluR2 and LimGluR2(R57A) was highly concentration-dependent with the largest effects between ~EC10-EC50 and the relation was shifted to higher concentrations for the R57A mutation, (Fig. 5H, I; Fig. S5G). Together these data suggest a supra-linear activation of mGluR2 by two agonists versus one.
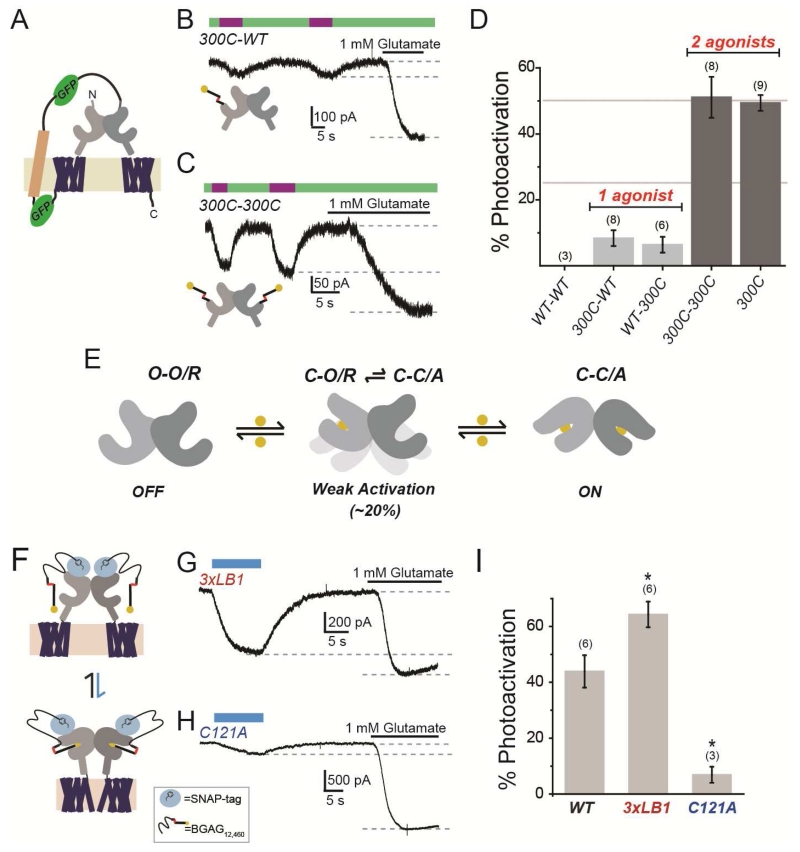
We next aimed to determine directly if agonist binding in only one subunit can activate mGluR2 and to quantify the relationship of ligand occupancy to activation. To clearly define the stoichiometry of label-able and unlabel-able subunits we adapted a tandem dimer approach used previously for microbial opsins (Kleinlogel et al., 2011), in which a transmembrane linker connects the C-terminus of a first copy of mGluR2 to the N-terminus of a second (Fig. 6A, S6A). The tandem dimer had the expected size on a western blot (Fig. S6A), expressed reasonably well (Fig. S6B), displayed normal apparent glutamate affinity (Fig. S6C), and showed photobleaching patterns consistent with the predicted assembly (indicating the presence of two GFPs within each complex) (Fig. S6D, E). We introduced the L300C MAG attachment site into the LBD of one subunit or both subunits within a tandem, labeled maximally with MAG and analyzed photoswitch efficiency. Tandem 300C-WT and WT-300C dimers showed weak, but clear photocurrents that were <10% in amplitude compared to the current evoked by 1 mM glutamate (Fig. 6B). We confirmed that these small currents were mGluR2-dependent through blockade with LY341495 (Fig. S6F). These photocurrents were not dependent on ambient glutamate in our bath solution because they persisted in the low affinity 300C(R57A) mutant (Fig. S6G). Consistent with the behavior described above for incomplete MAG labeling of homo-dimeric LimGluR2(L300C) (Fig. 5C), a low concentration (1 μM) of glutamate potentiated the photocurrent of 300C-WT (Fig. S6H). 300C-300C tandem dimers showed > 5-fold larger photoactivation than when only one subunit has a cysteine (300C-WT or WT-300C), and this level was the same as seen in homo-dimers formed by assembly of mGluR2-L300C monomers (Fig. 6C, D). 300C-300C tandem dimers also had faster photocurrents compared to 300C-WT tandem dimers (Fig. S6I), indicating that increased occupancy increases both the amplitude and speed of activation.
Figure 6. Photoactivation analysis of receptor cooperativity in tandem dimers and dimer interface mutants.
(A) Schematic of mGluR2 tandem dimer. Transmembrane linker contains two GFPs (green) and the transmembrane segment of the H+,K+ ATPase (beige).
(B-C) Representative traces showing that, compared to saturating glutamate, docking of glutamate of D-MAG-0 in a single subunit within a tandem dimer weakly activates mGluR2 (B), whereas docking in both subunits strongly activates (C). Tethering of D-MAG-0 to subunits is via introduced cysteine in one (300C-WT) or both (300C-300C) subunits.
(D) Summary of photoactivation relative to 1 mM glutamate for various conditions. Activation with 2 agonists is >5× as efficient as 1 agonist. All constructs are tandem dimers except for “300C,” which is the standard non-tandem LimGluR2 construct. The numbers of cells tested for each condition are shown in parentheses.
(E) Model of occupancy-dependent activation of mGluR2, where LBD is either open (O) or closed (C) and the receptor is either resting (R) or activated (A).
(F) Schematic of SNAP-mGluR2 photoactivation by BGAG12,460.
(G-H) Representative traces showing photoactivation of SNAP-3xLB1 (G) or SNAP-C121A (H).
(I) Summary of photoactivation relative to 1 mM glutamate for dimer interface mutants. The numbers of cells tested for each condition are shown in parentheses.
Error bars show S.E.M. calculated from multiple experiments (N.≥3).
Based on prior evidence that the activated state of the receptor has both LBDs closed and reoriented (CC/A) (Vafabakhsh et al, 2015), these findings suggest that one agonist weakly activates the receptor during rare spontaneous closures of the un-liganded subunit and that when two ligands are bound the receptor stably occupies this conformation and is fully activated (Fig. 6E).
We next sought to determine the role of the LBD interfaces in the cooperativity of mGluR2 activation by analyzing the photo-activation properties of the interface mutants with the largest effects on function, 3xLB1 and C121A. Because these mutants expressed poorly combined with the L300C substitution, we turned to an alternative approach for attachment of the photoswitchable glutamate ligand in which to an N-terminal SNAP domain is covalently labeled by benzylguanine-azobenzene-glutamate (“BGAG”) to enable photo-activation in cis (Fig. 6F) (Broichhagen et al, 2015), similar to what is observed with MAG. We used a BGAG variant, BGAG12,460, which, following maximal labeling, acts as a full agonist when bound, but provides partial activation due to incomplete photoisomerization (Broichhagen et al, 2015) and, thus, should produce a population with a mix of receptors where 0, 1, or 2 subunits are liganded within a dimer. Thus, photoswitch efficiency relative to saturating glutamate should provide a measure of the relative activation efficiency of partially liganded dimers. Consistent with smFRET experiments, SNAP-mGluR2(3xLB1) showed enhanced photoactivation while SNAP-mGluR2(C121A) showed a major decrease in photoactivation (Fig. 6G-IJ). These results argue that weakening of the hydrophobic LB1 interface promotes intersubunit cooperativity while removal of the intersubunit disulfide (C121A) weakens the ability of subunits to communicate and produce activation of receptors liganded in only one subunit.
mGluR2/3 heterodimers: conformational dynamics, basal activity, and cooperativity
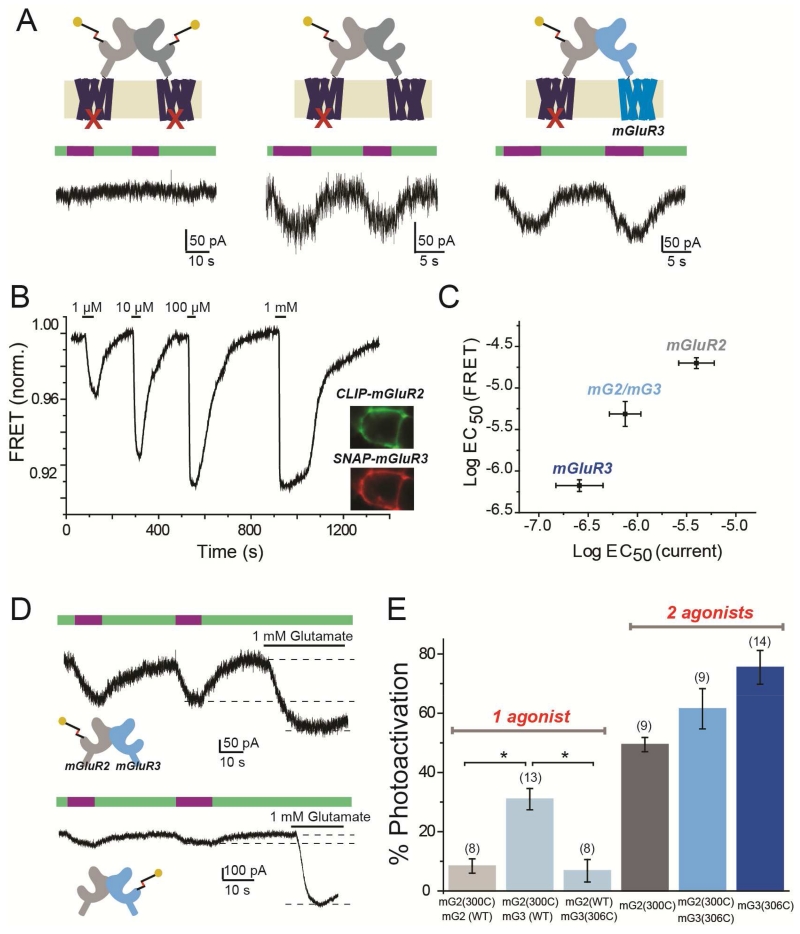
Since mGluR2 and mGluR3 readily heterodimerize in heterologous cells (Fig. 4, S4) and have overlapping native expression patterns (Ohishi et al., 1993a; Ohishi et al., 1993b; Petralia et al., 1996) that make them likely candidates for in vivo heterodimerization, we wondered if mGluR2/mGluR3 (“mG2/mG3”) heterodimers form functional glutamate receptors. Because there are no agonists that fully distinguish mGluR2 from mGluR3, we again turned to subunit selective activation with MAG. We first asked if LimGluR2 activation can cross-activate mGluR3. To focus our analysis on the freely assembled mG2/mG3 heterodimer, without interference from mGluR2 and mGluR3 homodimers that are formed in the same cells, we targeted mGluR2 for photo-control by MAG using LimGluR2 and also introduced the mutation F756D into the same subunit, to prevent G protein coupling (Francesconi and Duvoisin, 1998). This photo-activatable but non-functional LimGluR2(F756D) subunit was co-expressed with a non-photo-activatable (no MAG attachment site) but functional subunit to enable GIRK photocurrent to be generated only by the heterodimeric combination of the two. First, we confirmed the expected lack of photocurrent when LimGluR2(F756D) was expressed alone (Fig. 7A, left). When mGluR2WT was co-expressed with LimGluR2(F756D), photocurrent was observed, suggesting cross-activation to the intact G protein-coupling site of the WT subunit from the MAG-liganded LimGluR2(F756D) subunit (Fig. 7A, center). When mGluR3WT was co-expressed with LimGluR2(F756D), photocurrent was also observed (Fig 7A, right), suggesting that cross-activation also occurs between mGluR2 and mGluR3, setting the stage for experiments on heteromeric cooperative interaction.
Figure 7. mGluR2/mGluR3 heterodimers exhibit trans-activation, intermediate glutamate affinity, and assymmetric cooperativity.
(A) LimGluR2(F756D) G protein coupling mutant has no photocurrent (left), unless co-expressed with mGluR2WT (middle) or mGluR3WT (right).
(B) Representative trace showing glutamate-induced decreases in ensemble FRET between co-expressed CLIP-mGluR3 (labeled with donor) and SNAP-mGluR2 (labeled with acceptor) in a HEK293T cell. Inset shows cell donor (CLIP-mGluR3) and acceptor (SNAP-mGluR2) fluorescence images.
(C) Summary of glutamate EC50 determinations from measurement of GIRK activation (current) versus LBD conformational change (FRET) for mGluR2, mGluR3, and mGluR2/mGluR3 (“mG2/mG3”). FRET for mG2/mG3 obtained from co-expression, as in (B); GIRK current from tandem linked mGluR2-mGluR3 (“mG2-mG3”).
(D) GIRK current traces showing single-subunit photoactivation of linked mG2-mG3 heterodimers via photoactivation of only mGluR2 (top) or only mGluR3 (bottom).
(E) Summary of photoactivation (from left to right) with 1 subunit liganding of mGluR2 in mG2-mG2, in mG2-mG3, or 1 subunit liganding of mGluR3 in mG2-mG3, or 2 subunit labeling in unlinked mGluR2, linked mG2-mG3 or unlinked mGluR3. * indicates statistical significance (unpaired t-test, p=0.003 between mG2(300C)-mG2(WT) and mG2(300C)-mG3(WT); p=0.004 between mG2(300C)-mG3(WT) and mG2(WT)-mG3(306C). The numbers of cells tested for each condition are shown in parentheses.
Error bars show S.E.M. calculated from multiple experiments (N≥3).
We first asked how the ~10-fold higher glutamate affinity of mGluR3 (Conn and Pin, 1997); Vafabakhsh et al, 2015) would influence the behavior of mGluR2 in the mG2/mG3 heterodimer. To determine glutamate affinity, we measured ensemble inter-LBD FRET between an acceptor fluorophore on SNAP-mGluR2 and a donor fluorophore on CLIP-mGluR3 in HEK293T cells. Glutamate induced large FRET decreases, with an EC50 that was intermediate between that of homodimers of mGluR2 and mGluR3 (Fig. 7B, S7A). To confirm this functionally we turned to a measure of GIRK channel activation. We obtained a pure population of mG2/mG3 heterodimers through tandem heterodimers (“mG2-mG3”) which showed an intermediate apparent glutamate affinity for GIRK current activation (Fig. 7C, S7B,C). We next asked how ligand occupancy to mGluR2 or mGluR3 subunits within a heterodimer mediates receptor activation.
To directly measure subunit interaction between mGluR2 and mGluR3, we complemented photo-agonism of mGluR2 (“LimGluR2”), with photo-agonism of mGluR3 (i.e. LimGluR3: mGluR3(Q306C) + D-MAG-0). We previously observed enhanced photoactivation of LimGluR3 relative to LimGluR2 (Levitz et al, 2013), which made us wonder if mGluR2 cooperativity is altered in mG2/mG3 heterodimers. Strikingly, photoactivation of only mGluR2 within a mG2(300C)-mG3(WT) heterodimer elicited robust photocurrents of ~30% amplitude relative to saturating glutamate (Fig. 7D, E). This photoactivation was more than 3-fold larger than what was seen when only one mGluR2 subunit was activated in an mGluR2 homodimer (Fig. 6B, 7E). In contrast, photoactivation of only the mGluR3 subunit in an mG2(WT)-mG3(306C) heterodimer showed an efficiency of ~10%, similar to single-subunit activation in the mGluR2 homodimer (Fig. 7D, E). Thus, unexpectedly, activation of mGluR2/3 heterodimers is asymmetric. In addition, mG2(300C)-mG3(306C) heterodimers showed intermediate photoactivation relative to LimGluR2 or LimGluR3 (Fig. 7E, S7D). To probe the molecular mechanism of this asymmetric subunit cooperativity, we turned to inter-subunit FRET.
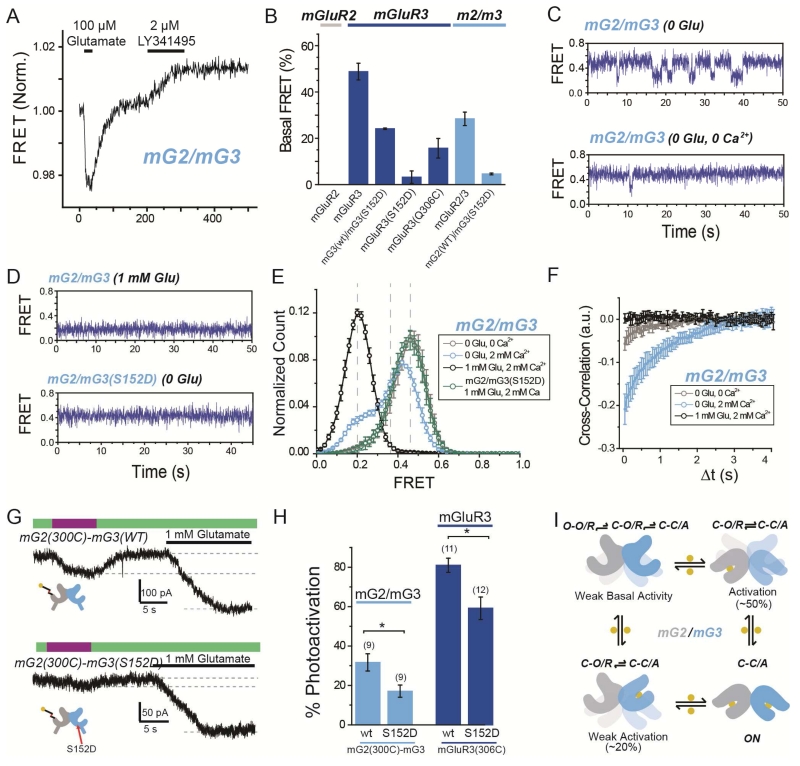
Whereas mGluR2 displays minimal spontaneous LBD dynamics in the absence of glutamate, mGluR3 shows rapid LBD dynamics, due primarily to partial agonism by Ca2+, which produce basal activity (Vafabakhsh et al., 2015). We asked whether the presence of mGluR3 in a mG2/mG3 heterodimer would confer basal dynamics by measuring basal FRET in the mG2/mG3 heterodimer in physiological (2 mM) Ca2+ and zero glutamate by measuring the increase in inter-subunit FRET that is induced by the competitive antagonist LY341495. Unlike in the mGluR2 homodimer, whose basal FRET was insensitive to LY341495, the basal FRET of the mG2/mG3 heterodimer was substantially reduced by LY341495 to ~60% of that seen in the mGluR3 homodimer (Fig. 8A,B). The mutation S152D, which targets a site implicated in Ca2+ binding (Kubo et al., 1998) and reduces basal dynamics in mGluR3 homodimers (Vafabaksh et al., 2015), also reduced basal FRET in the mG2/mG3 heterodimer (Fig. 8B). Interestingly, introduction of S152D into just one subunit of mGluR3 reduced basal FRET to a level similar to that seen in mG2/mG3 (Fig. 8B). These results support a model whereby Ca2+ binds to each subunit within a dimer to produce basal activity by inducing LBD closure.
Figure 8. Basal conformational dynamics of mGluR2/3 heterodimers provide a mechanism for enhanced single subunit activation.
(A) Representative ensemble mGluR2/mGluR3 (“mG2/mG3”) FRET trace shows an increase in FRET in response to the orthosteric antagonist LY341495 in the absence of glutamate.
(B) Summary of percentage of basal FRET that is LY341495-sensitive in mGluR2, mGluR3, and mG2/mG3 variants. Basal FRET in mGluR3 is reduced by mutation 306C and nearly-abolished by mutation S152D.
(C) smFRET traces for mG2/mG3 in the absence of glutamate and presence (top) or absence (bottom) of 2 mM Ca2+.
(D) smFRET traces for mG2/mG3 in the presence of saturating glutamate (top) or for mGluR2/3(S152D) in the absence of glutamate (bottom).
(E) Histogram showing smFRET distribution for mG2/mG3 and mG2/mG3(S152D) in various conditions.
(F) Cross-correlation analysis of mGluR2/3 reveals basal dynamics that are diminished by either the removal of Ca2+ or the addition of saturating glutamate.
(G) Representative GIRK current traces showing single-subunit photoactivation of mGluR2 in tandem heterodimer of mG2(300C)-mG3(WT) (top) or mG2(300C)-mG3(S152D) (bottom).
(H) Summary of effect on photoactivation of introduction of S152D mutation in mG2-mG3 tandem heterodimers (light blue) or mGluR3 homodimers (dark blue). * indicates statistical significance (unpaired t-test, p=0.003 between mGluR2/3 heterodimers and p=0.008 between mGluR3 homodimers). The numbers of cells tested for each condition are shown in parentheses.
(I) Conformational model of ligand occupancy-dependent activation of mGluR2/mGluR3 heterodimers.
Error bars show S.E.M. calculated from multiple experiments (N≥3).
To obtain a quantitative view of mG2/mG3 conformational dynamics we performed smFRET experiments following SimPull isolation. mG2/mG3 transitioned between three distinct FRET states, with peak values similar to those observed in mGluR2 and mGluR3 homodimers. In 2 mM Ca2+ and zero glutamate, mG2/mG3 displayed rapid dynamics with frequent visits to the active, low FRET state (Fig 8C, top). The occupancy of the low FRET state was ~20%, intermediate between what was seen in mGluR2 and mGluR3 homodimers and consistent with ensemble FRET results (Fig. 8B). Both mG2/mG3 basal dynamics and low FRET occupancy were reduced either by application of LY341495, removal of Ca2+, or introduction of the S152D mutation (Fig. 8C-F; Fig. S8A, B). Application of glutamate to mG2/mG3 heterodimers increased the occupancy of the low FRET state in a dose-dependent manner and decreased dynamics (Fig. S8A). We also applied DCG-IV to see if this would uncover differences in the stability of the active conformation. Compared to mGluR2, saturating DCG-IV in mG2/mG3 showed more complete occupancy of the low FRET state as well as decreased dynamics (Fig. S8C, D). Together these data show that mG2/mG3 heterodimers visit the same three conformations as their parent homodimers and possess properties of mGluR3, including Ca2+-dependent basal dynamics and a stabilized active state relative to mGluR2.
We hypothesized that the enhanced single subunit activation by mGluR2 within the mG2/mG3 heterodimer is due to spontaneous closure of the mGluR3 subunit, which enhances population of the low FRET (C-C/A) state when only mGluR2 is liganded. To test this we introduced the S152D mutation into the mGluR3 subunit and measured the ability of mGluR2 to activate the mG2(WT)/mG3(S152D) receptor via D-MAG-0. As expected, photoactivation of mG2(300C)-mG3(S152D) was weaker than in mG2(300C)-mG3(WT) (Fig. 8G, H). Consistent with a role for receptor dynamics in determining ligand efficacy, LimGluR3(S152D) also showed reduced photocurrent efficacy compared to LimGluR3 (Fig. 8H; Fig. S8E, FB). Together, these data support a role for basal conformational dynamics in determining ligand efficacy.
Discussion
Dimerization has long been viewed as a defining feature of class C GPCRs. However, the molecular determinants of dimerization have been only partly defined and the function of dimerization has remained opaque. We used single molecule subunit counting in the plasma membrane of living cells and in isolated receptors to identify determinants of mGluR assembly. We confirm earlier work (Doumazane et al., 2011) that, unlike the formation of tetramers or higher ordered complexes in the related GABAB receptor (Calebiro et al., 2013; Maurel et al., 2008) (Comps-Agrar et al., 2011), mGluRs from all three groups form strict homodimers, and show that this applies at both low and high expression densities. By comparing low density conditions in oocytes to high density conditions in HEK293T cell lysate, we found that dimerization is mediated by high affinity LBD interactions and lower affinity TMD interactions. In contrast to an earlier report (El Moustaine et al., 2012), the TMD interface alone, in the absence of the entire extracellular domain, permits the formation of stable dimers at high expression densities. Analogous interactions may be relevant to Class A and B GPCRs, whose oligomerization has been the subject of ongoing debate (Lambert and Javitch, 2014; Bouvier and Hebert, 2014). Interestingly, the recent crystal structure of the TMD of mGluR1 identified an interface between TM1 helices (Wu et al., 2014), while a structure of the TMD of mGluR5 was obtained as a monomer (Dore et al., 2014). In contrast, a recent cross-linking study suggested that activation reorients a TMD interface involving TM4, TM5 and TM6 (Xue et al., 2015) and other studies have indicated that mGluR2 can interact with the 5-HT2A serotonin receptor via TM4 (Gonzalez-Maeso et al., 2008; Moreno et al., 2012).
The LBD interface in assembly and function
We focused our analysis on LBD interfaces, which deletion analysis showed to play a dominant role in receptor dimerization. At low expression density, mutation of the intersubunit disulfide bridge (C121 in mGluR2) compromises dimerization without eliminating it, whereas mutation of the hydrophobic LB1 interface results in complete monomerization. Thus, the conserved disulfide bond does not ensure dimerization when other interfaces are sufficiently weakened and the LB1 interface accounts for the dominant role of the LBD in dimerization. Consistent with this, the mGluR2 LBD alone efficiently dimerizes with full-length mGluR2, as previously observed with mGluR1 (Beqollari and Kammermeier, 2010).
In contrast to the requirement of trafficking to the plasma membrane on heteromerization in GABABRs (Margeta-Mitrovic et al., 2000), we find that monomerized mGluRs do traffic to the cell surface. Based on these findings, as well as the observations that mGluR2 is stationary in the plasma membrane and that application of high concentrations of DTT do not dissociate dimers, it appears that dimerization of mGluRs likely occurs in the endoplasmic reticulum, and that receptor reaches the plasma membrane as a stable dimer.
We used single molecule analysis to determine that mGluR2 readily heterodimerizes with the other group II mGluR, mGluR3, as well as with the group III mGluR, mGluR7, but not with either of the group 1 mGluRs, mGluR1 and mGluR5, consistent with earlier work (Doumazane et al., 2011). When surface expression levels are matched, mGluR2 heterodimerizes with a preference for the same group (equally well with itself as with mGluR3) over group III. Chimeras between mGluR1 and mGluR2 revealed that determinants of heterodimer specificity exist in both the LBD and the TMD. Because of its dominant role in assembly, we focused on the role of LBD interactions in activation.
Strikingly, weakening of the LB1 dimer interface increased the apparent affinity of mGluR2 for both glutamate and the partial agonist DCG-IV and increased DCG-IV efficacy. These effects were accompanied by spontaneous activation rearrangements in the absence of glutamate and by two changes to in the presence of glutamate: increased active state occupancy and a startling shift to a lower FRET level than seen in wildtype. These effects suggest that the LB1 mutations result in a larger than normal activation motion that produces a more stable activated conformation. This suggests that the normal function of the LB1 interface is to stabilize the resting conformation and, thereby, prevent basal activity in absence of glutamate, while at the same time limiting the sensitivity to glutamate.
In contrast, mutation to prevent the formation of an intersubunit disulfide bond (C121A) had the opposite effect: it decreased the apparent affinity for agonists and decreased the stability of the active state. Moreover, this mutation also increased the FRET level of the resting state suggesting that the function of the intersubunit disulfide bond is to set the resting conformation and stabilize the active state. Interestingly, LB1 interface mutations in the background of C121A still introduced basal dynamics, but did not prevent the increase in basal FRET values or the destabilization of the active state, suggesting that these interfaces work in concert via distinct mechanisms.
Subunit cooperativity in mGluR activation
The importance to activation of the dimer interface is consistent with earlier evidence that a coordinated rearrangement of the subunits within a dimer takes place upon ligand binding (Kunishima et al., 2000) (Tateyama et al., 2004)(Doumazane et al., 2013). This raises a fundamental question about the activation mechanism: How does activation change when agonist binds to only one versus to both subunits in either a homodimer or heterodimer? Previous work that addressed this question in dimers composed of a wildtype subunit and a mutant subunit with a lower glutamate affinity showed that the wildtype subunit appeared to increase the affinity of the mutant subunit (Kammermeier and Yun, 2005; Kniazeff et al., 2004) (Tateyama and Kubo, 2011). However, there remained a concern that mutations of the orthosteric binding pocket that lowered affinity might have also altered activation in unforeseen ways. To overcome this complication, we turned from soluble ligands to the fast and reversible light-driven liganding of a specific LBD that is enabled by a “photoswitchable glutamate on a string,” D-MAG-0, that is anchored to a cysteine introduced into the LBD of a particular subunit. We showed D-MAG-0 to function as a high efficacy agonist, even in the low-affinity R57A, indicating that in its cis state its effective concentration is very high. Unique to a PTL, high ligand occupancy could be achieved and reversed rapidly but in only the fraction of subunits to which MAG is conjugated.
To precisely define ligand occupancy within a dimer, we developed a tandem dimer approach that allowed for the expression of heterodimeric mGluRs of defined stoichiometry. The ability to reversibly and repeatedly photoisomerize D-MAG-0, and the high signal-to-noise and reproducibility of GIRK current readout, allowed us to quantify activation even when it was weak. By targeting D-MAG-0 conjugation to one subunit within the linked dimer we found, definitively, that binding of agonist to one subunit is able to activate mGluR2. However, when two agonists were made to bind, we found that activation is ~5× larger, indicating substantial cooperativity.
Model of dimer interaction and cooperativity in mGluR activation
In order to reconcile our finding of non-linear occupancy-dependent activation with the 3 conformations observed in smFRET, we adapted earlier models of mGluR activation, in which the LBD is open (O) in absence of agonist and the receptor is in a resting intersubunit orientation (R) and the LBD is closed (C) when agonist binds, leading to a secondary reorientation rearrangement (A) that activates the receptor (Vafabakhsh et al., 2015)(Fig. 5E). In this updated model, spontaneous closure of the unliganded subunit is required for the receptor to enter the C-C/A state when only one ligand is bound. The ability of unliganded LBDs to close is consistent with our observation that, in the absence of glutamate, mGluR2 occasionally visits the medium FRET state. Furthermore, we hypothesize that closure of one LBD allosterically shifts the dynamic equilibrium of the unliganded subunit. This model can explain the biphasic dose response curve observed for a heterodimer between mGluR2WT and a subunit with diminished binding (“mGluR2-YADA”), when the response is effector activation (Kniazeff et al., 2004) or smFRET measurement of occupancy of the active, low FRET state (Vafabakhsh et al., 2015).
Cooperativity experiments with tandem mGluR2/mGluR3 (“mG2/mG3”) heterodimers were particularly illuminating for understanding activation cooperativity. Single subunit activation of the mGluR2, but not the mGluR3, subunit displayed efficient photoactivation compared to what was observed in mGluR2 homodimers. Consistent with this, smFRET analysis of mG2/mG3 conformational dynamics revealed mGluR3-like basal dynamics that could be diminished by the introduction of mutation S152D or removal of Ca2+. Supporting the notion of a central role for receptor dynamics in the cooperative response to ligands, introduction of S152D into mG2-mG3 tandem dimers reduced the activation via the mGluR2 subunit. The observed asymmetric activation cooperativity argues that the basal dynamics of mG2/mG3 are not distributed throughout the receptor, but are confined to the mGluR3 subunit. This suggests that Ca2+ binding occurs in a subunit autonomous manner to stabilize LBD closure rather than to directly induce dimer reorientation. Furthermore, S152 is located at the hinge between the upper and lower lobes of the LBD indicating that it may provide energy for LBD closure. Finally, experiments with the partial agonist DCG-IV revealed increased low FRET occupancy for mG2/mG3, implying that the active state is more energetically favorable in the mG2/mG3 heterodimer than in the mGluR2 homodimer.
Together these experiments allow us to propose a conformational model that describes the asymmetrical activation of mG2/mG3 heterodimers where the properties of the un-liganded subunit determine efficacy (Fig. 8I). mG2/mG3 may provide an advantageous combination of its two parent receptors by combining the basal activity of mGluR3, which may provide a useful inhibitory tone, with the non-linear response of mGluR2 (since physiologically the mGluR3 subunit is likely to bind glutamate first due to its higher affinity), which may sharpen the response to ligand compared to mGluR3. Its unique combination of basal activity, cooperativity, and glutamate affinity, indicates that mG2/mG3 may be uniquely tuned to distinct spatiotemporal synaptic glutamate profiles. In situ hybridization studies have revealed a significant overlap in expression of mGluR2 and mGluR3 in a number of brain regions and cell types, including golgi cells of the cerebellum, dentate gyrus granule cells, the olfactory bulb and many parts of the thalamus and cortex (Ohishi et al., 1993a); (Ohishi et al., 1993b; Petralia et al., 1996), suggesting that the cooperative gating that we describe is likely to be physiologically relevant. The finding that mGluR2 and 3 readily heterodimerize also provides an important cautionary note for the interpretation of pharmacological and immunohistochemical studies, which often struggle to specifically target mGluR2 and mGluR3.
Methods
Molecular Cloning and Gene Expression
All experiments were performed using rat isoforms of mGluRs. See Suppementary Information for more detail on specific clones.
Single Molecule Subunit Counting and Pulldown
Subunit counting in Xenopus oocytes was performed as previously described (Ulbrich and Isacoff, 2007). SiMPull of mGluRs was performed 24-48 hours after transfection of HEK293T cells as previously described (Vafabakhsh et al., 2015). See Supplemental Experimental Methods for more detail.
Ensemble and Single Molecule FRET
Ensemble and single molecule FRET experiments were performed as previously described (Vafabakhsh et al., 2015).
Whole Cell Patch Clamp and Two Electrode Voltage Clamp Electrophysiology
HEK293 cell electrophysiology was performed as previously described (Vafabakhsh et al., 2015). Drugs were applied using a gravity-driven perfusion system and illumination was controlled using a DG-4 (Sutter) in combination with excitation filters (379/34 and 500/24 nm for D-MAG-0, 445/20 nm for BGAG12,460). See Supplementary Experimental Methods for details on oocyte two-electrode voltage clamp electrophysiology. Data was analyzed with Clampfit (Molecular Devices) and Prism (Graph pad).
Supplementary Material
Highlights.
mGluRs assemble into strict homo- or hetero-dimers, primarily via inter-LBD interactions
LBD interfaces modulate conformational dynamics and activation
Homomeric subunit interaction between mGluR2 subunits yields activation cooperativity
Spontaneous dynamics yield assymetric cooperativity in mGluR2/3 heterodimers
Acknowledgements
We thank A. Reiner for helpful discussion, M. Kienzler, J. Broichhagen, A. Damijoinatas, and P. Leippe for chemical synthesis, and R. Arant, H. Okada, and C. Stanley for technical assistance. The work was supported by the National Institutes of Health Nanomedicine Center for the Optical Control of Biological Function (2PN2EY018241) and instrumentation award (S10 RR028971) and the National Science Foundation Major Research Instrumentation Award (1041078) and EAGER award (IOS-1451027).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.L. and E.Y.I designed experiments with input from all authors. J.L., C.H, S.B., Z.F, and R.V. performed experiments. J.L., C.H., and S.B analyzed data. J.L. and E.Y.I wrote the manuscript.
References
- Beqollari D, Kammermeier PJ. Venus fly trap domain of mGluR1 functions as a dominant negative against group I mGluR signaling. J Neurophysiol. 2010;104:439–448. doi: 10.1152/jn.00799.2009. [DOI] [PubMed] [Google Scholar]
- Bouvier M, Hebert TE. CrossTalk proposal: Weighing the evidence for Class A GPCR dimers, the evidence favours dimers. J Physiol. 2014;592:2439–2441. doi: 10.1113/jphysiol.2014.272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J, Damijoinatas A, Levitz J, Sokol K, Leippe P, Konrad D, Isacoff EY, Trauner D. Orthogonal optical control of a class C G protein-coupled receptor using a SNAP-tethered photoswitch. ACS Central Science. 2015;1:383–393. doi: 10.1021/acscentsci.5b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D, Rieken F, Wagner J, Sungkaworn T, Zabel U, Borzi A, Cocucci E, Zurn A, Lohse MJ. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci U S A. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EC, Berlin S, Levitz J, Kienzler MA, Yuan Z, Madsen D, Larsen DS, Isacoff EY. Two-photon brightness of azobenzene photoswitches designed for glutamate receptor optogenetics. Proc Natl Acad Sci U S A. 2015;112:E776–785. doi: 10.1073/pnas.1416942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comps-Agrar L, Kniazeff J, Norskov-Lauritsen L, Maurel D, Gassmann M, Gregor N, Prezeau L, Bettler B, Durroux T, Trinquet E, et al. The oligomeric state sets GABA(B) receptor signalling efficacy. EMBO J. 2011;30:2336–2349. doi: 10.1038/emboj.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- Delille HK, Becker JM, Burkhardt S, Bleher B, Terstappen GC, Schmidt M, Meyer AH, Unger L, Marek GJ, Mezler M. Heterocomplex formation of 5-HT2A-mGlu2 and its relevance for cellular signaling cascades. Neuropharmacology. 2012;62:2184–2191. doi: 10.1016/j.neuropharm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Fabre L, Zwier JM, Trinquet E, Pin JP, Rondard P. Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2013;110:E1416–1425. doi: 10.1073/pnas.1215615110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, Pin JP. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 2011;25:66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- El Moustaine D, Granier S, Doumazane E, Scholler P, Rahmeh R, Bron P, Mouillac B, Baneres JL, Rondard P, Pin JP. Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc Natl Acad Sci U S A. 2012;109:16342–16347. doi: 10.1073/pnas.1205838109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre S, Casado V, Devi LA, Filizola M, Jockers R, Lohse MJ, Milligan G, Pin JP, Guitart X. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi A, Duvoisin RM. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J Biol Chem. 1998;273:5615–5624. doi: 10.1074/jbc.273.10.5615. [DOI] [PubMed] [Google Scholar]
- Geng Y, Bush M, Mosyak L, Wang F, Fan QR. Structural mechanism of ligand activation in human GABA(B) receptor. Nature. 2013;504:254–259. doi: 10.1038/nature12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Maeso J, Ang RL, Yuen T, Chan P, Weisstaub NV, Lopez-Gimenez JF, Zhou M, Okawa Y, Callado LF, Milligan G, et al. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature. 2008;452:93–97. doi: 10.1038/nature06612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S, Deupi X, Opitz C, Heydenreich FM, Tsai CJ, Brueckner F, Schertler GF, Veprintsev DB, Grzesiek S. Backbone NMR reveals allosteric signal transduction networks in the beta1-adrenergic receptor. Nature. 2016;530:237–241. doi: 10.1038/nature16577. [DOI] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ, Yun J. Activation of metabotropic glutamate receptor 1 dimers requires glutamate binding in both subunits. J Pharmacol Exp Ther. 2005;312:502–508. doi: 10.1124/jpet.104.073155. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Wilkins AD, Lichtarge O, Wensel TG. Determinants of endogenous ligand specificity divergence among metabotropic glutamate receptors. J Biol Chem. 2015;290:2870–2878. doi: 10.1074/jbc.M114.622233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinlogel S, Terpitz U, Legrum B, Gokbuget D, Boyden ES, Bamann C, Wood PG, Bamberg E. A gene-fusion strategy for stoichiometric and co-localized expression of light-gated membrane proteins. Nat Methods. 2011;8:1083–1088. doi: 10.1038/nmeth.1766. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Bessis AS, Maurel D, Ansanay H, Prezeau L, Pin JP. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol. 2004;11:706–713. doi: 10.1038/nsmb794. [DOI] [PubMed] [Google Scholar]
- Kubo Y, Miyashita T, Murata Y. Structural basis for a Ca2+-sensing function of the metabotropic glutamate receptors. Science. 1998;279:1722–1725. doi: 10.1126/science.279.5357.1722. [DOI] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- Lambert NA, Javitch JA. CrossTalk opposing view: Weighing the evidence for class A GPCR dimers, the jury is still out. J Physiol. 2014;592:2443–2445. doi: 10.1113/jphysiol.2014.272997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz J, Pantoja C, Gaub B, Janovjak H, Reiner A, Hoagland A, Schoppik D, Kane B, Stawski P, Schier AF, et al. Optical control of metabotropic glutamate receptors. Nat Neurosci. 2013;16:507–516. doi: 10.1038/nn.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Tsai MC, Davenport CM, Smith CM, Veit J, Wilson NM, Adesnik H, Kramer RH. A Comprehensive Optogenetic Pharmacology Toolkit for In Vivo Control of GABAA Receptors and Synaptic Inhibition. Neuron. 2015;88:879–891. doi: 10.1016/j.neuron.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ. Dimerization in GPCR mobility and signaling. Curr Opin Pharmacol. 2010;10:53–58. doi: 10.1016/j.coph.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Knoflach F, Broger C, Ohresser S, Kratzeisen C, Adam G, Stadler H, Kemp JA, Mutel V. Identification of essential residues involved in the glutamate binding pocket of the group II metabotropic glutamate receptor. Mol Pharmacol. 2001;60:944–954. doi: 10.1124/mol.60.5.944. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, Hilger D, Lerch MT, Kobilka TS, Thian FS, Hubbell WL, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- Maurel D, Comps-Agrar L, Brock C, Rives ML, Bourrier E, Ayoub MA, Bazin H, Tinel N, Durroux T, Prezeau L, et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JL, Muguruza C, Umali A, Mortillo S, Holloway T, Pilar-Cuellar F, Mocci G, Seto J, Callado LF, Neve RL, et al. Identification of three residues essential for 5-hydroxytryptamine 2A-metabotropic glutamate 2 (5-HT2A.mGlu2) receptor heteromerization and its psychoactive behavioral function. J Biol Chem. 2012;287:44301–44319. doi: 10.1074/jbc.M112.413161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2007;104:3759–3764. doi: 10.1073/pnas.0611577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajo K, Ulbrich MH, Kubo Y, Isacoff EY. Stoichiometry of the KCNQ1 - KCNE1 ion channel complex. Proc Natl Acad Sci U S A. 2010;107:18862–18867. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi H, Shigemoto R, Nakanishi S, Mizuno N. Distribution of the messenger RNA for a metabotropic glutamate receptor, mGluR2, in the central nervous system of the rat. Neuroscience. 1993a;53:1009–1018. doi: 10.1016/0306-4522(93)90485-x. [DOI] [PubMed] [Google Scholar]
- Ohishi H, Shigemoto R, Nakanishi S, Mizuno N. Distribution of the mRNA for a metabotropic glutamate receptor (mGluR3) in the rat brain: an in situ hybridization study. J Comp Neurol. 1993b;335:252–266. doi: 10.1002/cne.903350209. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Niedzielski AS, Wenthold RJ. The metabotropic glutamate receptors, mGluR2 and mGluR3, show unique postsynaptic, presynaptic and glial localizations. Neuroscience. 1996;71:949–976. doi: 10.1016/0306-4522(95)00533-1. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Hauschild BC. Cys-140 is critical for metabotropic glutamate receptor-1 dimerization. J Biol Chem. 2000;275:34245–34251. doi: 10.1074/jbc.M005581200. [DOI] [PubMed] [Google Scholar]
- Reiner A, Arant RJ, Isacoff EY. Assembly stoichiometry of the GluK2/GluK5 kainate receptor complex. Cell Rep. 2012;1:234–240. doi: 10.1016/j.celrep.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Levitz J, Isacoff EY. Controlling ionotropic and metabotropic glutamate receptors with light: principles and potential. Curr Opin Pharmacol. 2015;20:135–143. doi: 10.1016/j.coph.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C, Miller JK, Hyrc K, Dikranian S, Mennerick S, Takeuchi Y, Goldberg MP, O’Malley KL. Covalent and noncovalent interactions mediate metabotropic glutamate receptor mGlu5 dimerization. Mol Pharmacol. 2001;59:46–53. [PubMed] [Google Scholar]
- Romano C, Yang WL, O’Malley KL. Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J Biol Chem. 1996;271:28612–28616. doi: 10.1074/jbc.271.45.28612. [DOI] [PubMed] [Google Scholar]
- Sounier R, Mas C, Steyaert J, Laeremans T, Manglik A, Huang W, Kobilka BK, Demene H, Granier S. Propagation of conformational changes during mu-opioid receptor activation. Nature. 2015;524:375–378. doi: 10.1038/nature14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Moriyoshi E, Tsuchiya D, Jingami H. Negative cooperativity of glutamate binding in the dimeric metabotropic glutamate receptor subtype 1. J Biol Chem. 2004;279:35526–35534. doi: 10.1074/jbc.M404831200. [DOI] [PubMed] [Google Scholar]
- Tateyama M, Abe H, Nakata H, Saito O, Kubo Y. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1alpha. Nat Struct Mol Biol. 2004;11:637–642. doi: 10.1038/nsmb770. [DOI] [PubMed] [Google Scholar]
- Tateyama M, Kubo Y. The intra-molecular activation mechanisms of the dimeric metabotropic glutamate receptor 1 differ depending on the type of G proteins. Neuropharmacology. 2011;61:832–841. doi: 10.1016/j.neuropharm.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+ Proc Natl Acad Sci U S A. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Shimada Y, Takeshita T, Kajimura N, Nomura S, Sekiyama N, Otomo J, Usukura J, Nakanishi S, Jingami H. Cryptic dimer interface and domain organization of the extracellular region of metabotropic glutamate receptor subtype 1. J Biol Chem. 2000;275:28144–28151. doi: 10.1074/jbc.M003226200. [DOI] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- Vischer HF, Castro M, Pin JP. G Protein-Coupled Receptor Multimers: A Question Still Open Despite the Use of Novel Approaches. Mol Pharmacol. 2015;88:561–571. doi: 10.1124/mol.115.099440. [DOI] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Rovira X, Scholler P, Zhao H, Liu J, Pin JP, Rondard P. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat Chem Biol. 2015;11:134–140. doi: 10.1038/nchembio.1711. [DOI] [PubMed] [Google Scholar]
- Yin S, Noetzel MJ, Johnson KA, Zamorano R, Jalan-Sakrikar N, Gregory KJ, Conn PJ, Niswender CM. Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J Neurosci. 2014;34:79–94. doi: 10.1523/JNEUROSCI.1129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.