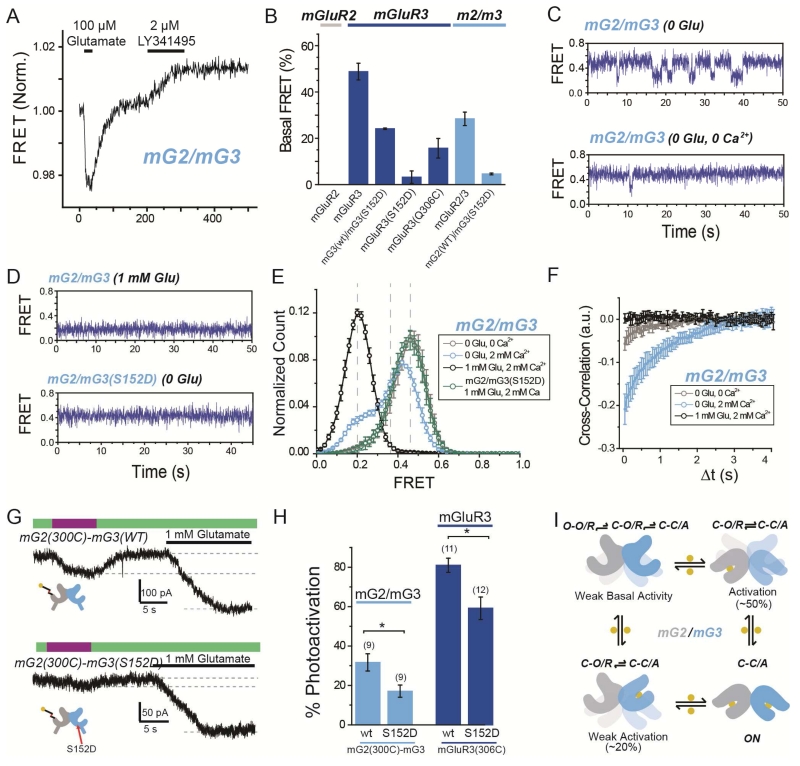
Figure 8. Basal conformational dynamics of mGluR2/3 heterodimers provide a mechanism for enhanced single subunit activation.
(A) Representative ensemble mGluR2/mGluR3 (“mG2/mG3”) FRET trace shows an increase in FRET in response to the orthosteric antagonist LY341495 in the absence of glutamate.
(B) Summary of percentage of basal FRET that is LY341495-sensitive in mGluR2, mGluR3, and mG2/mG3 variants. Basal FRET in mGluR3 is reduced by mutation 306C and nearly-abolished by mutation S152D.
(C) smFRET traces for mG2/mG3 in the absence of glutamate and presence (top) or absence (bottom) of 2 mM Ca2+.
(D) smFRET traces for mG2/mG3 in the presence of saturating glutamate (top) or for mGluR2/3(S152D) in the absence of glutamate (bottom).
(E) Histogram showing smFRET distribution for mG2/mG3 and mG2/mG3(S152D) in various conditions.
(F) Cross-correlation analysis of mGluR2/3 reveals basal dynamics that are diminished by either the removal of Ca2+ or the addition of saturating glutamate.
(G) Representative GIRK current traces showing single-subunit photoactivation of mGluR2 in tandem heterodimer of mG2(300C)-mG3(WT) (top) or mG2(300C)-mG3(S152D) (bottom).
(H) Summary of effect on photoactivation of introduction of S152D mutation in mG2-mG3 tandem heterodimers (light blue) or mGluR3 homodimers (dark blue). * indicates statistical significance (unpaired t-test, p=0.003 between mGluR2/3 heterodimers and p=0.008 between mGluR3 homodimers). The numbers of cells tested for each condition are shown in parentheses.
(I) Conformational model of ligand occupancy-dependent activation of mGluR2/mGluR3 heterodimers.
Error bars show S.E.M. calculated from multiple experiments (N≥3).