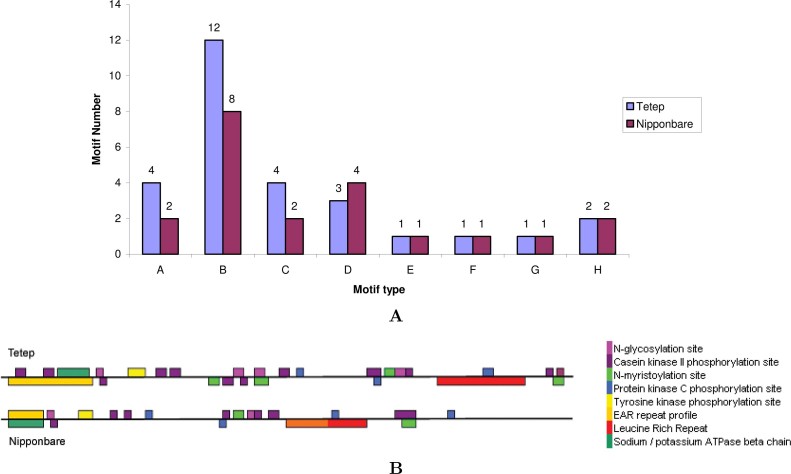
Fig. 5.
Analysis of motifs and their physical positions in the Pi-kh gene isolated from indica (Tetep) and japonica (Nipponbare) sequences. A. The number of motifs predicted in Pi-kh alleles of indica and japonica. A: N-glucosylation site; B: casein kinase II phosphorylation site; C: N-myristorylation site; D: protein kinase C phosphorylation site; E:tyrosine kinase phosphorylation site; F: EAR repeat profile; G: Na/K-ATPase β-chain; H: leuecine-rich repeat. B. Physical positions of motifs distributed in Pi-kh alleles of indica and japonica sequences. The types of motifs are shown in different colors.