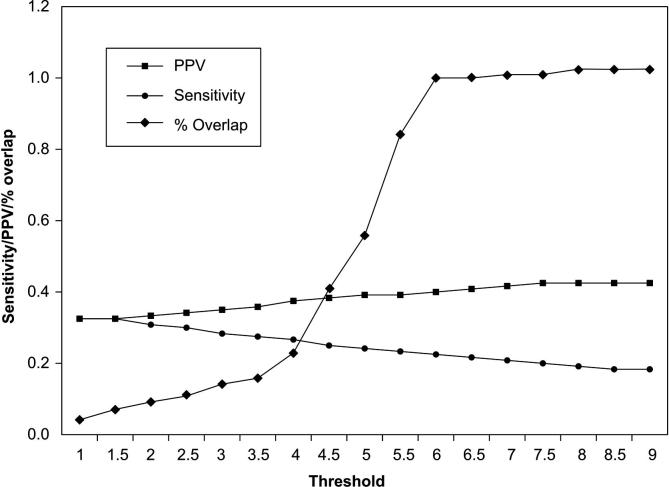
Figure 1.
Sensitivity, PPV and percentage of detections that overlap with known ncRNA genes for ncRNAscout on a shuffled E. coli genome Half of the shuffled genome was used as training data and the other half was used as test data. ncRNAscout demonstrated the best performance at a threshold of 6.0 with a PPV of 0.393, sensitivity of 0.213, and percentage of overlap with known ncRNA genes of 1.0.