Abstract
Atovaquone is a hydroxy-naphthoquinone that is used to treat parasitic and fungal infections including Plasmodium falciparum (malaria), Pneumocystis jivorecii (pneumonia) and Toxoplasma gondii (toxoplasmosis). It blocks mitochondrial oxidation of ubiquinol in these organisms by binding to the ubiquinol oxidation site of the cytochrome bc1 complex. Failure of atovaquone treatment has been linked to the appearance of mutations in the mitochondrially encoded gene for cytochrome b. In order to determine the optimal parameters required for inhibition of respiration in parasites and pathogenic fungi and overcome drug resistance, we have synthesized and tested the inhibitory activity of novel hydroxy-naphthoquinones against blood stage P. falciparum and liver stage P. berghei and against cytochrome bc1 complexes isolated from yeast strains bearing mutations in cytochrome b associated with resistance in Plasmodium, Pneumocystis, and Toxoplasma. One of the new inhibitors is highly effective against an atovaquone resistant Plasmodium and illustrates the type of modification to the hydroxy-naphthoquinone ring of atovaquone that might mitigate drug resistance.
Keywords: Malaria, Plasmodium, Pneumocystis, Toxoplasma, Atovaquone, Hydroxy-naphthoquinones, Cytochrome bc1 complex
1. Introduction
Atovaquone (ATV, Fig. 1 A) is a hydroxy-naphthoquinone (NQ) used to prevent and treat infections caused by Plasmodium falciparum (malaria) and Pneumocystis (pneumonia), a major cause of illness and death in immunocompromised individuals. Ubiquinol oxidation is blocked upon atovaquone binding to the ubiquinol oxidation site of the cytochrome bc1 complex, which inhibits respiration in these parasites. Although the bc1 complex is essential for energy transduction in all eukaryotic cells, recent studies indicate that during the erythrocytic stages of growth the essential function of the bc1 complex in P. falciparum is to maintain ubiquinone oxidized and thus available as an electron acceptor for dihydroorotate dehydrogenase [1]. The latter enzyme is essential for pyrimidine biosynthesis in the parasite.
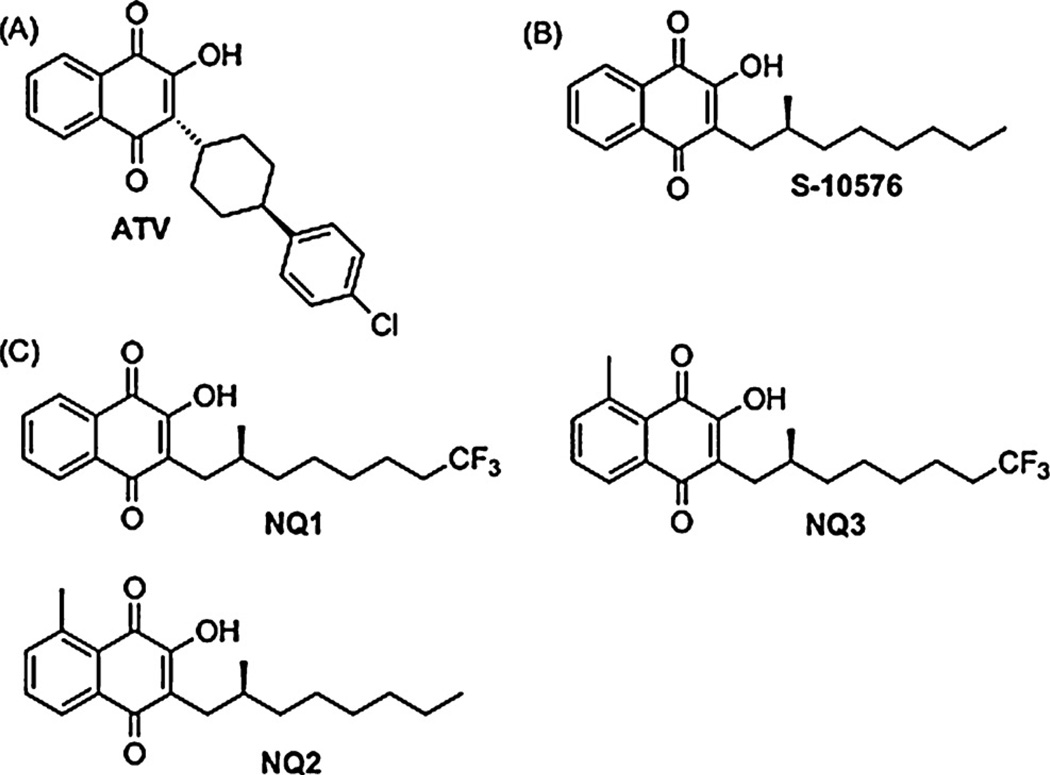
Fig. 1.
(A) Atovaquone (B) S-10576 (C) 2-OH-3-(2-methyl-trifluorooctyl)-naphthoquinone (NQ1), 2-OH-3-(2-methyl-octyl)-8-methyl-naphthoquinone (NQ2) (NQ3), 2-OH-3-(2-methyl-trifluorooctyl)-8-methyl-naphthoquinone.
Since the bc1 complex is essential for ubiquinol reoxidation, and thus indirectly essential for pyrimidine biosynthesis in Plasmodium, it is an attractive target for development of novel anti-malarials. This has led to the design of several new bc1 inhibitors, such as the 4-pyridone derivatives [2,3] including GW844520 [4], and several novel hydroxy-naphthoquinones [5], all of which bind to the ubiquinol oxidation site in the bc1 complex and are effective at low nM concentrations against the parasite.
Malarone® is a drug combination containing atovaquone and proguanil, an inhibitor of dihydrofolate reductase, which is also essential for pyridimine biosynthesis in the parasite. Spontaneously arising mutations conferring resistance to atovaquone have been linked to the mitochondrially encoded gene for cytochrome b [6], and reports of Malarone® treatment failure describe mutations at codon 268 of the cytochrome b gene that result in substitution of Tyr with Asn, Ser or Cys in cytochrome b in P. falciparum [7].
Since atovaquone also inhibits the yeast cytochrome bc1 complex, we have previously developed Saccharomyces cerevisiae as a model to study resistance conferring cytochrome b mutations such as Y279S (which corresponds to Y268S in Plasmodium), L275F (L248F in Pneumocystis numbering) and M139L (which corresponds to M129L in T. gondii) [8–11]. Using this model system we were able to show that the mutations at Tyr-268 in Plasmodium disrupt non-covalent interactions between atovaquone and the binding pocket in the bc1 complex [8], and that the most commonly observed mutation in atovaquone resistant Pneumocystis, L248F, creates an unfavorable bulge in the binding pocket [9]. These structural changes explain how these mutations impair the efficacy of the drug.
We have recently synthesized the stereochemically active derivative of a potent cytochrome bc1 complex inhibitor S-10576 (Fig. 1B), featuring a metabolically stable trifluoromethyl function and amethyl substituted aromatic ring (Fig. 1C and Refs 12, 13). The racemic mixture of R- and S-10576 had first been tested by Fieser and coworkers as an anti-malarial [14], who showed that although it was effective against avian malaria, it was ineffective in humans. It was then shown that R/S-10576 is excreted as a water-soluble derivative in which the terminal methyl group is removed from the 2-methyl-octyl side chain by oxidative decarboxylation to yield a 2-methyl-heptyl carboxylic acid side chain [15]. We reasoned that a trifluoromethyl group at the terminus of the side-chain should block this detoxification reaction.
The 8-methyl group was introduced into the naphthoquinone ring in NQ2 and NQ3 on the basis of molecular modeling that indicated it should be possible to create non-covalent interaction between the ligand and the Rieske iron-sulfur protein by this modification. We expected that interaction with this nuclear encoded protein would lessen the frequency of spontaneously arising resistance conferring mutations that otherwise are directed to weakening interactions between the mitochondrial encoded cytochrome b, which is more prone to spontaneously arising mutations than a nuclear gene.
Here we report the activity of these hydroxy-naphthoquinones (“NQ’s”) against the malaria parasite and cytochrome bc1 complexes isolated from wild-type yeast, and yeast carrying Y279S, L275F and M139L cytochrome b mutations in order to determine the parameters required for inhibition of drug resistant parasites and fungi.
2. Experimental procedures
2.1. Materials
Atovaquone was a gift from GlaxoSmithKline. The hydroxy-naphthoquinones S-10576 (13) and the NQ1-3 [16] were synthesized as previously described. The NQ’s were dissolved in dimethyl sulfoxide at 2 mM concentration and stored at −20 °C.
2.2. Isolation of yeast mitochondrial membranes
The yeast strains with wild-type background and cytochrome b mutations L275F, Y279S and M139L were grown in 2% YPD media as detailed by Kessl et al. [8,11,17]. Mitochondrial membranes were prepared by breaking yeast cells with glass beads in a bead beater as described elsewhere [18] with the following modifications. Before breaking the cells, they were suspended in buffer volume equivalent to two times the wet weight of the cell pellet and 1 mM diisopropylfluorophosphate was added to the buffer. An equal volume of glass beads (Sigma, 425–600 µM) was then added and the cells were broken in a glass bead beater and membranes recovered by centrifugation [18]. After washing the membranes with 50 mM Tris, pH 8.0, the membranes were stored at −20 °C in 50 mM Tris, pH 8.0, containing 50% glycerol. Quantification of the bc1 complex was performed spectrophotometrically [19], using extinction coefficients of 17.5 mM−1 cm−1 at 553–539 for cytochrome c1 [20] and 25.6 mM−1 cm−1 at 563–579 for the average absorbance of the bH and bL hemes in cytochrome b [21].
2.3. Ubiquinol-cytochrome c reductase activity measurements
Cytochrome c reductase activities of membrane preparations were assayed in 50 mM potassium phosphate, pH 7.0, 250 mM sucrose, 2 mM EDTA, 1 mM NaN3, 2.5 mM KCN, 0.01% dodecylmaltoside, and 40 µM cytochrome c at 23 °C. The mitochondrial membranes were diluted to a cytochrome bc1 complex concentration of approximately 5 nM in the assay buffer, inhibitor was added to the assay mixture and allowed to stir with the enzyme for 1 min, after which the reaction was started by adding 50 µM 2,3-dimethoxy-5-methyl-6-decyl-1,4-benzoquinol, an analogue of ubiquinol. Reduction of cytochrome c was monitored in an Aminco DW-2a spectrophotometer at 550 versus 539 nm in dual wave-length mode. Data were collected and analyzed using an Online Instrument Systems Inc. computer interface and software. IC50 values were determined from titration curves of inhibitor concentration versus activity which were analyzed with the Origin 5.0 program (OriginLab Corp.) The data points were fitted to a monophasic or biphasic curve, according to whichever gave the better fit.
2.4. In vitro activity of inhibitors against P. falciparum blood stages and P. berghei liver stages
The in vitro activities of the test compounds against intraerythrocytic stages of P. falciparum strains W2, D6, TM91C235 and TM90C2B were evaluated using the traditional radio-labeled hypoxanthine assay using the method of Desjardins et al. [22], as modified by Milhous et al. [23]. These strains represent different relative degrees of resistance to atovaquone and other malaria drugs. The laboratory clone W2 (Indochina) is chloroquine resistant, whereas the D6 (Sierra-Leone) clone is chloroquine sensitive but naturally less susceptible to mefloquine. The laboratory adapted field isolate TM91C235 (Thailand) is resistant to mefloquine, chloroquine and pyrimethamine. W2, D6 and TM91C235 are susceptible to atovaquone. TM90C2B is a laboratory adapted field isolate from Thailand that is atovaquone resistant. Two different cell cultures of TM90C2B of the same origin strain were used in the susceptibility assay; one was established and cryopreserved in 1995 and the other in 2007.
The in vitro activities of the test compounds against exoerythrocytic stages of P. berghei were evaluated using a luminescence-based “inhibition of liver stage development assay” (or ILSDA), based on the method described by Sacci [24]. Briefly, white 96-well microtiter plates previously coated with ECL Cell Attachment Matrix (20 µg/ml final, Millipore) were seeded with 2.5 × 104 HepG2 cells [a human hepatocellular carcinoma cell line, American Tissue Culture Collection (ATCC)] per well in 100 µl culture medium and incubated overnight in a humidified atmosphere at 37C in 5% CO2-air. Liver cells were cultured in ATCC-formulated Eagle’s Minimum Essential Medium (EMEM) with Earle’s salts supplemented with 10% fetal bovine serum, 0.19% (w/v) sodium bicarbonate, 1.36 mg/ml bovine serum albumin, 0.68 mg/ml insulin, 2 mM l-glutamine, and 1 mM non-essential amino acids. After overnight incubation, the culture medium was removed from the liver cells and replaced with 50 µl of 1 × 104 luciferase-expressing P. berghei sporozoites in liver cell medium per well. Freshly isolated sporozoites from day 21 infected Anopheles stephensi mosquito salivary glands were harvested using a modification of the Ozaki method [25] and used for all experiments. The luciferase-expressing parasites were previously generated from a stably transformed P. berghei clone (ANKA-507m6cl1) that were kindly provided by Drs. C.J. Janse and A.P. Waters [26] through the Malaria Research and Reference Reagent Resource Center (MR4). The sporozoite-HepG2 cocultures were then incubated for 3 h at 37 °C in 5% CO2–air to allow for hepatocellular sporozoite invasion. After the incubation, extracellular sporozoites are removed by washing with liver cell medium plus 200 U/ml penicillin–streptomycin and 0.05 mg/ml gentamycin. Next, 100 µl of liver cell medium plus antibiotics containing drug or test compound (in 9 two-fold serial dilutions) was transferred to the plates and incubated 48 h at 37 °C in 5% CO2–air. After 48 h, 10 µl of 1× d-luciferin (200× stock at 30 mg/ml, Caliper Life Sciences) and further incubated at 37 °C for 30 min. Luminescence was read using the Packard TopCount NXT microplate luminescence counter (Packard BioScience Co.) and examined for relative luminescence units (RLUs) per well. The drug concentrations (x values) were transformed using the formula X = log[X] and plotted against the RLUs (y values). The data were then analyzed with Prism 4.0 (GraphPad Software, Inc., San Diego, CA) by nonlinear regression (sigmoidal dose–response/variable slope equation) to yield drug 50% inhibitory concentrations (IC50s).
In vitro toxicity was assessed using a standard MTT (3-(4,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide) assay, as modified from Caridha et al. [27]. Briefly, 96-well microtiter plates were seeded with 2.5 × 104 cells/well in 170 µl liver cell culture medium 24 h before the start of the assay. Serial dilutions of drug and test compounds (30 µl) were then incubated with the liver cells for 48 h.MTT (30 µl, 150 µg/ml final concentration) was added to each well. Plates were then incubated for 1 h followed by the removal of unincorporated MTT and air drying. The remaining purple formazan crystals were dissolved by adding 63 µl acidified isopropanol (95% isopropanol; 5% 2 N HCl). The absorbance was measured on a Tecan Infinite F200 plate reader at 540 nm. The data were then analyzed by nonlinear regression to yield IC50s, as described above.
2.5. Homology modeling
Sequences for P. falciparum proteins were obtained from PlasmoDB.org (NC 002375 and PF14 0373). Sequence alignments and homology models were created with Swiss-model version 8.05. The S. cerevisiae bc1 complex crystal structure was selected as the template for the modeling (PDB 3CX5) [28]. Structure models were validated using PROCHECK [29] and the overall stereochemical quality of the protein was assessed by Ramchandran plot analysis. The structures were visualized using Swiss-PDB Viewer version 4.0.
3. Results
3.1. Effect of inhibitors on P. falciparum growth
The hydroxy-naphthoquinones were tested for their ability to inhibit growth of several strains of P. falciparum in human red blood cells, including two atovaquone resistant isolates. The strains tested were D6, which is chloroquine sensitive, W2, which is chloroquine resistant, TM91C235, which is resistant to mefloquine, chloroquine and pyrimethamine, and two C2B isolates, which are atovaquone-resistant due to a Y268S mutation on cytochrome b (equivalent to Y279 in the yeast numbering), in the ubiquinol oxidation pocket at center P [30].
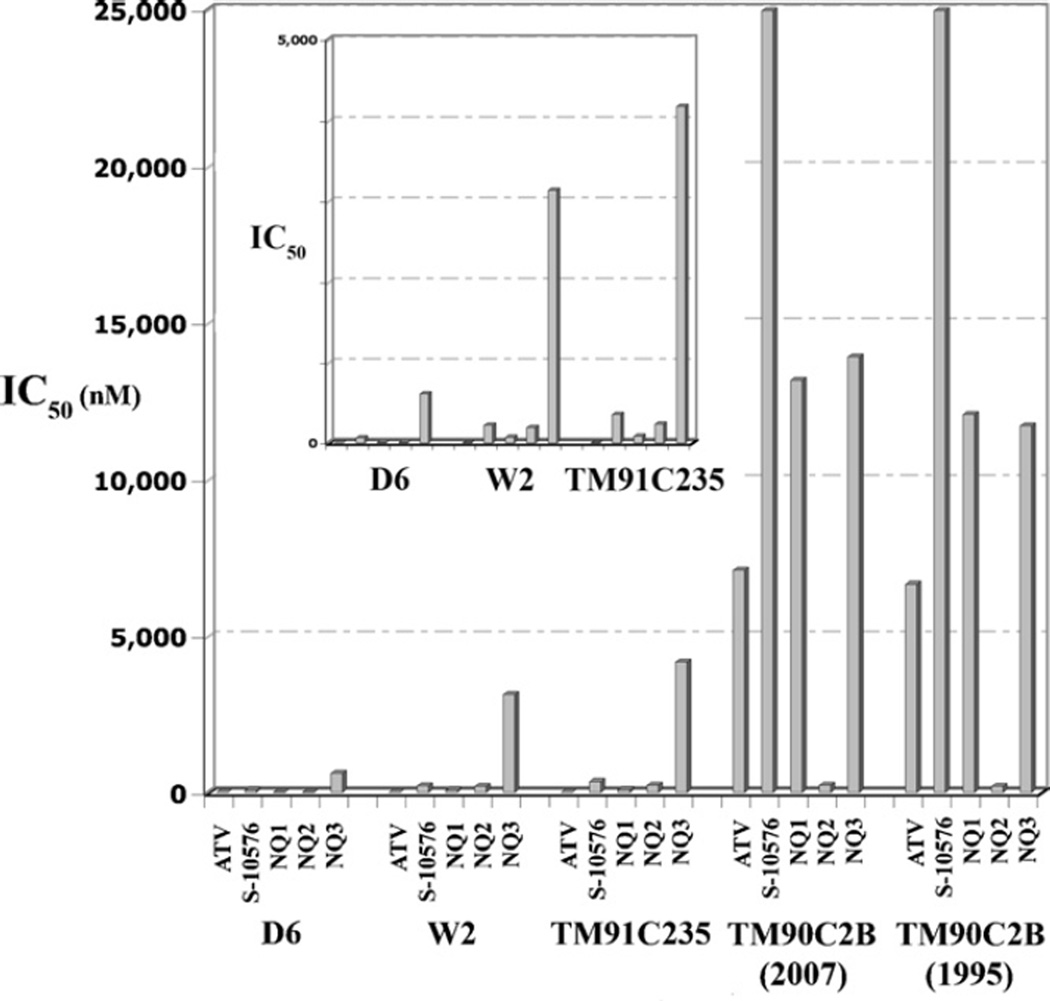
As shown in Fig. 2, all of the new hydroxy-naphthoquinone inhibitors were effective against the three Plasmodium strains that were not atovaquone resistant, D6, W2 and TM91C235, but none were as effective as atovaquone. The IC50 values for atovaquone against D6, W2 and TM91C235 were 0.34, 1.0 and 1.2 nM, respectively, while the values against those strains for S-10576 were 63, 214 and 348 nM. Introduction of a trifluoro group onto the side-chain of S-10576 resulted in improved efficacy against these strains in comparison to S-10576, with IC50 values for NQ1 of 5, 65, and 84 nM. However, the introduction of the 8-methyl group on the ring decreased the efficacy, resulting in IC50 values of 73, 191 and 229 nM for NQ2. The combination of the trifluoro group on the side-chain and the 8-methyl group on the ring resulted in an even greater loss of efficacy. Consequently, NQ3 was the least effective of the new inhibitors against D6, W2 and TM91C235, with IC50 values that ranged from 607 nM to 4.2 µM.
Fig. 2.
Effect of 2-hydroxy-naphthoquinones on growth of P. falciparum strains. The bar graphs show the IC50 values for inhibition of growth of different P. falciparum strains. The inset shows the results for the D6, W2 and TM91C25 strains on an expanded scale.
The C2B isolates are very resistant to atovaquone, as indicated by the increase in IC50 values by more than three orders of magnitude to 6.6–7.1 µM (Fig. 2). However, atovaquone was still more effective against the atovaquone resistant C2B isolates than the new hydroxy-naphthoquinone inhibitors, with the exception of NQ2. The IC50 values for NQ2 against the two C2B isolates were 193 and 234 nM, which are more than an order of magnitude lower than the IC50 values for atovaquone. Notably, the IC50 values for NQ2 against the atovaquone resistant isolates are comparable to the IC50 values against the atovaquone sensitive D6, W2 and TM91C235 strains, indicating that the resistance conferring mutation in the C2B parasites is ineffective against this inhibitor. The improved efficacy of NQ2 against the atovaquone resistant isolates validates the rationale of introducing the 8-methyl group onto the naphthoquinone ring.
We also tested the efficacy of the inhibitors against parasite sporozoites in hepatocytes and at the same time tested them for cytotoxicity against the hepatocytes alone. As shown in Table 1, S-10576 and NQ1 were approximately as effective as atovaquone in this assay and neither of these inhibitors was very toxic to the liver cells. Although NQ2 was also effective against the liver stage parasites, it exhibited a higher degree of hepatocyte toxicity.
Table 1.
Activity against sporozoites in hepatocytes.
| Inhibitor | IC50 against P. berghei (nM) |
Hepatocyte counterscreen (nM) |
Selectivity indexa |
| Atovaquone | 19.7 | 38,436.3 | 1953 |
| S-10576 | 96.2 | >66,580 | NT |
| NQ1 | 17.07 | 6826.4 | 399 |
| NQ2 | 246.5 | 2483.9 | 10.1 |
| NQ3 | 4451.8 | >54,290 | NT |
Selectivity index was calculated by dividing the IC50 value obtained against hepatocytes by the IC50 value for inhibition of P. berghei liver stage development.
NT= ‘Not toxic’ in this assay.
3.2. Effect of cytochrome b mutations on inhibition of the bc1 cytochrome complex by hydroxy-naphthoquinones
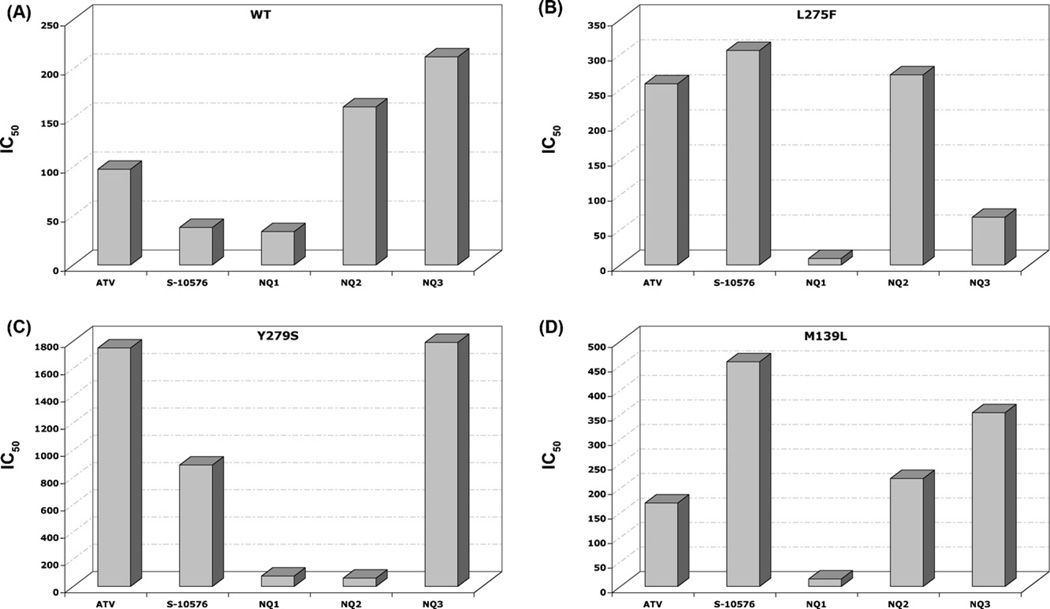
In order to assess the efficacy of the S-10576 analogs against target enzyme bearing cytochrome b mutations linked with atovaquone resistance in P. falciparum (L275F), Pneumocystis (Y279S), and T. gondii (M139L) mitochondrial membranes were prepared from wild-type yeast and yeast strains with the atovaquone-resistance conferring cytochrome b mutations. The cytochrome c reductase activities of the membranes were then measured in the presence of increasing concentrations of inhibitor. IC50 values calculated from the titration curves are shown in Fig. 3. It can be seen that the Y279S mutation confers the greatest degree of resistance to atovaquone, and it is thus not surprising that this is the most prevalent resistance-conferring mutation in P. falciparum [8,9].
Fig. 3.
Inhibitor efficacy of 2-hydroxy-naphthoquinones on bc1 activity of mitochondrial membranes isolated from (A) wild-type yeast and from (B) L275F (C) Y279S and (D) M139L yeast cytochrome b mutants. The cytochrome c reductase activity of mitochondrial membranes was measured as described in Section 2 in the presence of increasing concentrations of inhibitor. IC50 values were calculated from the titration curves and are expressed as nM concentration on bar graphs for each of the indicated inhibitors. The assays were performed in duplicate and the results are shown as averages.
Interestingly, as judged by IC50 values, NQ2 is more than two orders of magnitude more effective than atovaquone against the yeast enzyme with the Y279S mutation, as it was against the C2B Plasmodium strain (Fig. 2), which carries the equivalent mutation. However, in the in vitro assay NQ2 activity against the Y279S enzyme was biphasic and did not reach complete inhibition even at 500 nM (Supplemental Fig. S-1). Possible explanations for this biphasic titration are discussed below. Whereas S-10576 and the trifluorinated derivative NQ1 are equally effective and the most potent inhibitors of the enzyme from wild-type yeast, NQ1 is the most effective inhibitor of the enzymes with the L275F and M139L mutations and one of the two most effective against the Y279S mutation.
3.3. Relative efficacy of hydroxy-naphthoquinones against wild-type and atovaquone resistant cytochrome bc1 complexes
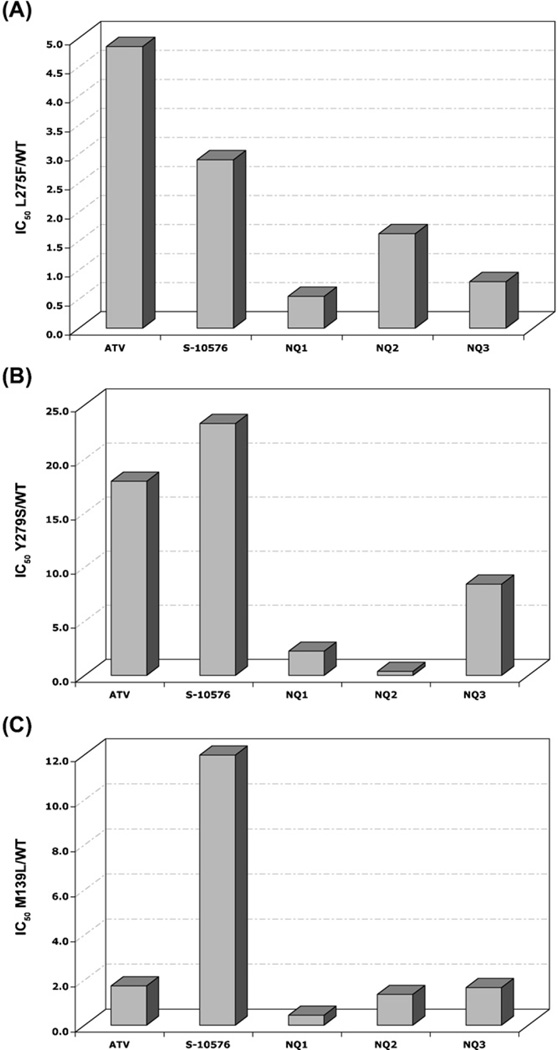
In order to provide an indication of the sensitivity of the mutant enzymes compared to the enzyme from the parent strain the IC50 values obtained against the mutant enzymes were divided by the IC50 value determined for the wild-type enzyme. The relative efficacy of the NQ’s against bc1 activities of mitochondrial membranes from the L275F, Y279S, and M139L mutants and from wild-type yeast are shown in Fig. 4. The yeast enzyme with the Y279S mutation, equivalent to the most prevalent atovaquone resistance-conferring mutation in Plasmodium, is more resistant than the wild-type enzyme to all of the inhibitors except NQ2, as shown by relative efficacy values > 1.0. The effectiveness of NQ2 against the Y279S enzyme is actually greater than against the enzyme from wild-type yeast, indicated by a relative efficacy value of 0.37, and as shown above (Fig. 3c), NQ1 is significantly more effective than atovaquone against the Y279S enzyme (Fig. 4B).
Fig. 4.
Relative efficacy of hydroxy-naphthoquinone inhibitors against (A) L275F (B) Y279S and (C) M139L yeast cytochrome bmutants. The bar graphs show the relative efficacy of the inhibitors determined by dividing the IC50 of the inhibitor against the mutant enzyme by the IC50 of the wild-type enzyme.
Notably, for each of the enzymes with mutations that confer resistance to atovaquone, NQ1 and NQ2 have lower relative efficacy values than atovaquone and S-10576, indicating these two inhibitors are more effective than the current therapeutic against the enzymes with resistance-conferring mutations in cytochrome b. Interestingly, the relative efficacy of NQ1 against the enzymes with the L275F (Fig. 4A) and M139L (Fig. 4C) mutations is < 1.0, indicating that this hydroxy-naphthoquinone more effectively inhibited the mutated cytochrome bc1 complexes than the wild-type enzyme. In other words, these two mutations that confer resistance to atovaquone make the bc1 complex more sensitive to NQ1 than the enzyme without the atovaquone resistance-conferring mutations.
3.4. Homology modeling of the atovaquone binding site of Plasmodium falciparum
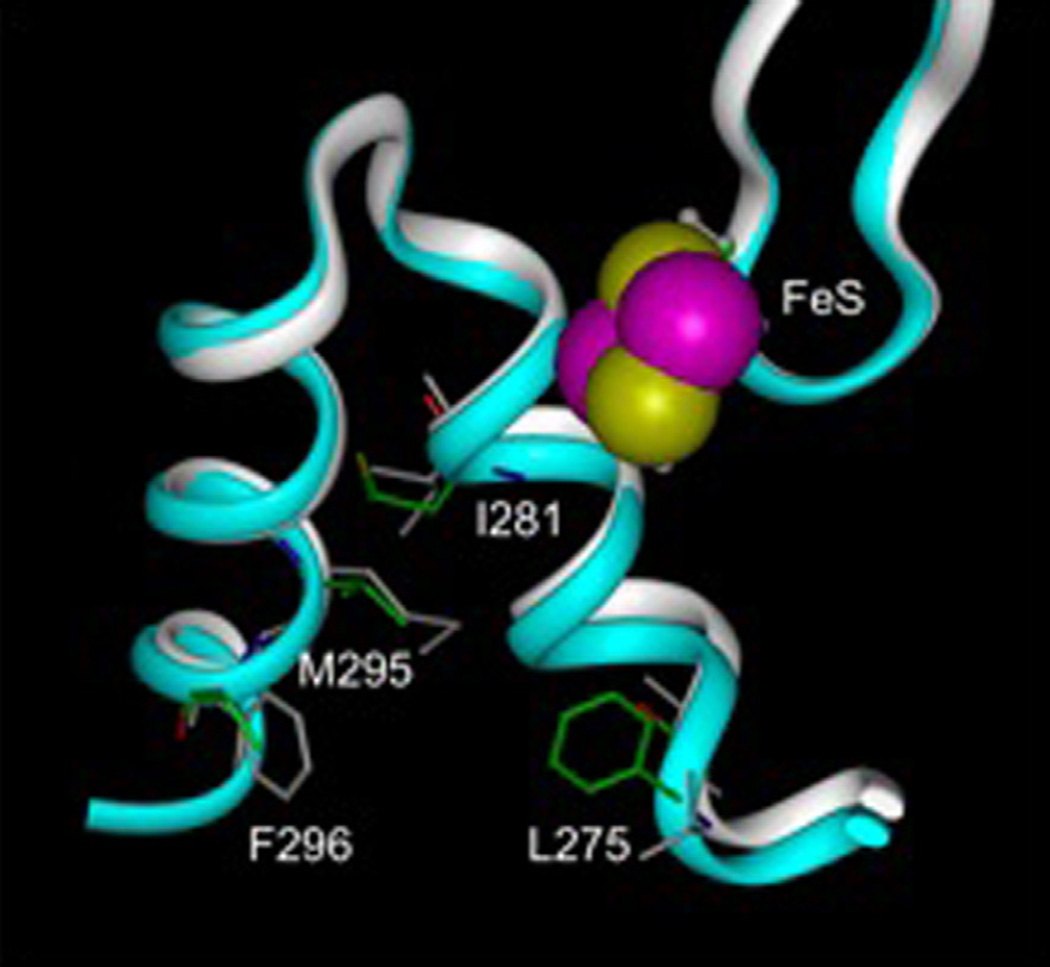
We used the crystal structure of the cytochrome bc1 complex from Saccharomyces cerevisiae (3CX5) as a template to generate homology models of P. falciparum cytochrome b and Rieske Fe-S protein. These models facilitated comparison of the structural differences of the atovaquone binding sites of the yeast and mutant parasite bc1 complexes as shown in Fig. 5, including non-conservative residue changes such as L275 to F, F296 to L, M295 to V and I281 to M that might account for any differences in inhibitor efficacies between the yeast and Plasmodium enzymes.
Fig. 5.
Overlaid models of center P of the bc1 complex from yeast and P. falciparum. Residues that are different between the two structures are shown as sticks and labeled. The yeast ribbon structure is shown in white and individual residues are gray. The ribbon of the P. falciparum model is colored blue and the carbon atoms of selected residues are colored green, oxygen atoms are red, nitrogen atoms are blue and hydrogen atoms are white. Van der Waals radii of the iron and sulfur atoms of the Rieske iron–sulfur cluster are colored purple and yellow, respectively.
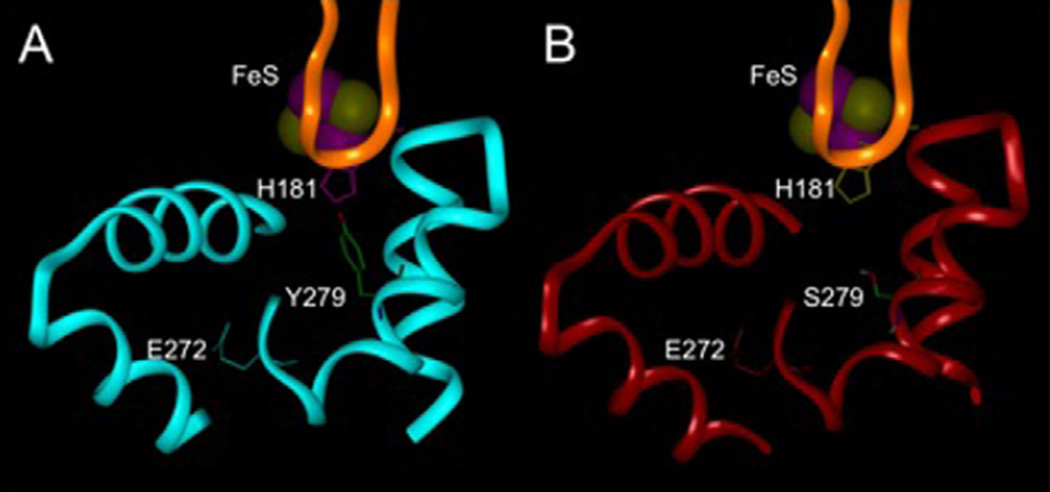
Previous modeling studies of yeast bc1 complexes have shown that when atovaquone is bound to the ubiquinol oxidation pocket at center P of the bc1 complex, the benzene ring of the NQ is part of an aromatic cluster composed of His-181 of the Rieske Fe-S protein opposite Tyr-279 of cytochrome b [9]. Substitution of Tyr-279 with serine decreases hydrophobic interactions between the binding site and atovaquone, which weakens the binding of the drug and accounts for the resistance. Fig. 6 shows this mutational change modeled in the enzyme from P. falciparum. It can be seen that the proximity of His-181 and the aromatic Tyr-279 form a relatively tight pocket abutting the cd1 helix in the enzyme from the wild-type parasite, as compared to the relatively open pocket with the aliphatic hydroxyl-containing Ser-279 in the mutant P. falciparum.
Fig. 6.
Molecular modeling of the atovaquone binding pocket of wild-type P. falciparum (A) and mutant Y279S (Y268S in Plasmodium numbering) (B). A portion of cytochrome b including the cd1 and ef helices, the ef loop, and part of helix C is shown in blue for the wild-type and red for the mutant. A portion of the Rieske iron-sulfur protein is shown in gold. The iron-sulfur cluster is at the top with the iron and sulfur atoms colored purple and yellow respectively. The carbon atoms of residue 279 are colored green, oxygen atoms are red, nitrogen atoms are blue and hydrogen atoms are white.
4. Discussion
Spontaneously arising mutations in cytochrome b confer resistance toward atovaquone, a hydroxy-naphthoquinone that is used to treat Plasmodium, Pneumocystis, and Toxoplasma infections. We aim to design new drugs that act in a manner similar to atovaquone but are effective against atovaquone resistant pathogens and are not rendered ineffective due to detoxification and excretion. Toward this end we have synthesized and tested the inhibitory activity of three novel hydroxy-naphthoquinones. To mitigate the effect of resistance conferring mutations that might arise in the mitochondrial encoded cytochrome b subunit that forms part of the atovaquone-binding pocket, we introduced a methyl group onto the naphthoquinone ring. Molecular modeling studies had indicated that this would result in non-covalent interactions with the Rieske iron–sulfur protein, a nuclear encoded protein less prone to spontaneously arising mutations than cytochrome b. To block the oxidative modification that was previously shown to render the chiral R/S-10576 ineffective against malaria in humans [15], we introduced a trifluoromethyl group onto the end of the side-chain in S-10576, which previous studies had shown was the active isomer in the chiral mixture [16]. We then tested these new hydroxy-naphthoquinones, along with atovaquone and S-10576, against malaria parasites growing in human red blood cells and hepatocytes and against cytochrome bc1 complexes isolated from yeast strains bearing mutations in cytochrome b associated with atovaquone resistance in Plasmodium, Pneumocystis, and Toxoplasma.
The most significant finding to emerge from testing the new naphthoquinones against P. falciparum is that the rationale of introducing amethyl group into the naphthoquinone ring to counteract resistance-conferring properties of the cytochrome b mutations was validated. The basis of this rationale was that molecular modeling studies indicated that the 8-methyl group would introduce a van der Waals interaction with Cys-180 of the yeast Rieske protein. This cysteine forms a disulfide bridge that is conserved in the Rieske protein across species, and when it was disrupted by mutagenesis, the Rieske protein was assembled into the yeast bc1 complex but was non-functional [31]. We thus expected that any potential resistance-conferring mutations that would abrogate the interaction of the 8-methyl group with the equivalent cysteine in Plasmodium would be lethal to the parasite. In addition, the Rieske protein is encoded by a nuclear gene and thus less susceptible to resistance conferring mutations which arise at much higher frequency in the mitochondrial encoded gene for cytochrome b, which is responsible for most of the non-covalent interactions with atovaquone. NQ2 was 2–3 orders of magnitude more effective than atovaquone against the atovaquone resistant C2B strains (Fig. 2) and also more effective than atovaquone against the yeast bc1 complex that carried the Y279S mutation (Figs. 3c and 4b) analogous to that in the C2B parasite. It was disappointing, however, that addition of a trifluoromethyl group to the side-chain in NQ3 abrogated the efficacy seen with NQ2, both in Plasmodium (Fig. 2) and in the yeast bc1 complex with the Y279S mutation (Fig. 3c). It should be noted, however, that the trifluoromethyl group is sterically similar to an isopropyl group. Thus, adding a trifluoromethyl group to the end of a 2-methyl-heptyl side-chain, instead of the 2-methyl-octyl side-chain as in NQ3, may improve the efficacy of this analogue. However, the intended protection against detoxification by adding the trifluoromethyl group may not be necessary, since NQ2 did retain significant activity against sporozoites in hepatocytes (Table 1). Further studies are required in order to address the metabolic stability of these potential anti-malarials.
In previous studies we used the yeast cytochrome bc1 complex as a surrogate to model the interaction of NQ’s with an atovaquone resistant bc1 complex of P. falciparum. When atovaquone is bound to the ubiquinol oxidation pocket of the bc1 complex Tyr-279 (Y268 in Plasmodium) forms part of a stable aromatic cluster that includes His-181 of the Rieske protein and the quinoid group of atovaquone [9]. Molecular modeling of the structural changes resulting from the Y279 mutations in the yeast cytochrome b revealed that atovaquone is ineffective against the Y279S/C cytochrome b mutants because the stabilizing effect of the aromatic cluster is lost and the nucleophilic serine and cysteine cannot form stabilizing hydrophobic interactions with the naphthoquinone ring. In the present study the increased efficacy of NQ1 and NQ2 against the Y279S yeast mutant suggests that the altered side-chains in these two compounds decrease the importance of the Y279 aromatic cluster in ligand binding.
Similar studies aimed at elucidating the basis for atovaquone resistance in Pneumocystis showed that replacing Leu-275 in the yeast cytochrome b with a phenylalanine generates an unfavorable bulge in the atovaquone-binding pocket that sterically collides with the chlorophenyl group of atovaquone, rendering the ligand unable to effectively bind to the pocket [11]. The increased efficacy of NQ1 against the yeast bc1 complex with the L275F mutation (Fig. 3B) may be due to the fact that the 2-methyloctyl side chain in NQ1 is smaller in volume than the cyclic side chains of atovaquone. This effect is lost when an 8-methyl group is added to the naphthoquinone ring in NQ2 and NQ3, probably due to the ligand moving deeper into the binding pocket in order to accommodate the increased size of the ring.
The slightly electropositive aromatic hydrogen atoms of atovaquone are thought to interact with the large electronegative sulfur atom of Met-139 [17]. In the case of the yeast enzyme with the M139L mutation (Fig. 3D), the affinity of atovaquone is decreased since the sulfur-aromatic link is replaced by a less favorable hydrophobic interaction. It is thus interesting that NQ1 is a more potent inhibitor of this mutant than atovaquone, although it is not obvious why, since both NQ’s are expected to interact similarly with Leu-139. As with the L275F enzyme the efficacy toward the M139L enzyme seen with NQ1 is lost when an 8-methyl group is added to the naphthoquinone ring in NQ2 and NQ3.
Interestingly, the trifluoromethyl derivatives NQ1 and NQ3 were more potent inhibitors of the activity of the L275F cytochrome bc1 complex (Fig. 3) than the parent compound S-10576. The Phe-275 residue is one of four amino acids predicted to be within 4 Å of the terminal groups of the alkyl chains of these NQ’s. The enhanced activity of NQ1 and NQ3 may be due to improved intermolecular interactions, such as van der Waals forces, between the terminal trifluoromethyl group of the ligands and Phe-275.
The current work illustrates both the usefulness and the limitations of the yeast bc1 complex as a surrogate to model the interactions of drugs such as atovaquone with their parasite target. The crystal structure of the yeast enzyme was clearly advantageous for modeling the molecular basis of atovaquone resistance resulting from mutations in cytochrome b [8,9,11,17]. In addition, screening of the hydroxy-naphthoquinones against the yeast enzyme with the L275F mutation correctly predicted that NQ2 is more effective than atovaquone against the drug resistant C2B malaria strain, although the improved efficacy is greater in the parasite than was seen with the isolated yeast enzyme. However, the relative efficacy of the other hydroxy-naphthoquinones against the L275F yeast enzyme did not extrapolate to the drug resistant parasite. The most notable discrepancy between the yeast in vitro testing and activity against the parasite was observed with the NQ1 inhibitor. Whereas the efficacy criteria showed this inhibitor to be generally the most effective against both normal and atovaquone resistant bc1 complexes in yeast (Figs. 3 and 4), it was relatively ineffective against the atovaquone resistant parasite (Fig. 2).
It was noted that whereas the very low IC50 value of NQ2 against the bc1 complex with the Y279S mutation (Fig. 3) correlated well with the efficacy of this inhibitor against the parasite (Fig. 2), it did not fully inhibit the activity of the enzyme even at concentrations >500 nM. This biphasic titration behavior was seen with all of the inhibitors against the enzymes with the atovaquone resistant mutations. One possible explanation is that this may be due to efficacy of the inhibitors being dependent on the redox status of the Rieske iron–sulfur protein, as was seen with undecyl-hydroxy-benzoxythiazole [32], which has a similar structure and binds in a similar manner to the quinol oxidation pocket as the hydroxy-naphthoquinones [33]. If the redox poise of the iron-sulfur protein is more reduced in vivo than in the isolated enzyme in the cytochrome c reductase assay, the in vitro assays will underestimate the efficacy of the inhibitors against the pathogen target. Another possible explanation is that the hydroxynaphthoquinone inhibitors bind to the ubiquinol oxidation pocket in the bc1 dimer in an anti-cooperative manner, with binding to one monomer being significantly tighter than to the other monomer. This behavior was previously observed with stigmatellin binding [34], and might be expected to occur to various extents with different NQ’s and with mutations that differently affect the structure of the binding pocket.
Another example of the limitations of the yeast enzyme as a model for the parasite target can be seen in the molecular modeling based on the yeast crystal structure. Molecular modeling of the hydroxy-naphthoquinones with the crystal structure of the yeast enzyme indicated that the addition of a trifluoromethyl group in NQ1 and NQ3 would be accommodated in the atovaquone binding pocket of the yeast bc1 complex, which it was, since the IC50 values for NQ1 and NQ3 against the enzyme from wild-type yeast were not significantly changed compared to those of the non-fluorinated S-10576 and NQ2, respectively (Fig. 3a). It was thus disappointing that the addition of the trifluoromethyl group in NQ3 markedly abrogated the efficacy toward Plasmodium that was observed with NQ2 (Fig. 2), as was also observed with the yeast enzyme with the Y279S mutation (Fig. 3c). Taken together these results suggest that it would be advantageous to modify the drug binding pocket of the yeast bc1 complex so that it more accurately mimics that of Plasmodium. A new method for introducing mutations into the cytochrome b gene may be especially useful in that regard [35].
Despite reports of atovaquone treatment failure, NQ’s are still potentially promising anti-malarial agents since Malarone is very well tolerated, needs to be taken for a relatively short period [36], and the mechanism of action is well known. Our findings demonstrate that minor modifications to the hydroxy-naphthoquinone ring structure may significantly mitigate the drug resistance that has compromised the effectiveness of atovaquone.
Supplementary Material
Acknowledgments
Supported by NIH Grant GM 20379. We thank Dr. Raul Covian for analysis of the inhibitor titration data. We are also grateful to the following individuals for conducting in vitro assays at the Walter Reed Army Institute of Research: Lucia Gerena and Norma Roncal executed the P. falciparum inhibitor susceptibility assay; Erin Harris conducted the inhibition of P. berghei liver stage development assay; and Dr. Diana Caridha performed the MTT hepatocyte cytotoxicity assay.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molbiopara.2011.01.002.
References
- 1.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 2.Yeates CL, Batchelor JF, Capon EC, Cheeseman NJ, Bueno JM, Chicharro J, Fernandez E, Fiandor JM, Gargallo-Viola D, de las Heras FG, Herreros E, Leon ML. Synthesis and structure-activity relationships of 4-pyridones as potential anti-malarials. J Med Chem. 2008;51:2845–2852. doi: 10.1021/jm0705760. [DOI] [PubMed] [Google Scholar]
- 3.Biagini GA, Fisher N, Berry N, Stocks PA, Meunieer B, Williams DP, Bonar-Law R, Bray PG, Owen A, O’Neill PM, Ward SA. Acridinediones: selective and potent inhibitors of the malaria parasite mitochondrial bc1 complex. Mol Pharmacol. 2008;73:1347–1355. doi: 10.1124/mol.108.045120. [DOI] [PubMed] [Google Scholar]
- 4.Xiang H, McSurdy-Freed J, Moorthy GS, Hugger E, Bambal R, Hau C, Ferrer S, Gargallo D, Davis CB. Preclinical drug metabolism and pharmacokinetic evaluation of GW844520, a novel anti-malarial mitochondrial electron transport inhibitor. J Pharm Sci. 2006;95:2657–2672. doi: 10.1002/jps.20681. [DOI] [PubMed] [Google Scholar]
- 5.Kongkathip N, Pradidphol N, Hasitapan K, Grigg R, Kao W-C, Hunte C, Fisher N, Warman AJ, Biagini GA, Kongsaeree P, Chuawong P, Kongkathip B. Transforming rhinacanthin analogues from potent anticancer agents into potent antimalarial agents. J Med Chem. 2010;53:1211–1221. doi: 10.1021/jm901545z. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999;33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 7.Sutherland CJ, Laundy M, Price N, Burke M, Fivelman QL, Pasvol G, Klein JL, Chiodini PL. Mutations in the Plasmodium falciparum cytochrome b gene are associated with delayed parasite recrudescence in malaria patients treated with atovaquone-proguanil. Malar J. 2008;7:240–261. doi: 10.1186/1475-2875-7-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessl JJ, Ha KH, Merritt AK, Lange BB, Hill P, Meunier B, Meshnick SR, Trumpower BL. Cytochrome b mutations that modify the ubiquinol-binding pocket of the cytochrome bc1 complex and confer anti-malarial drug resistance in Saccharomyces cerevisiae. J Biol Chem. 2005;280:17142–17148. doi: 10.1074/jbc.M500388200. [DOI] [PubMed] [Google Scholar]
- 9.Kessl JJ, Meshnick S, Trumpower BL. Modeling the molecular basis of atovaquone resistance in parasites and pathogenic fungi. Trends Parasitol. 2007;23:494–501. doi: 10.1016/j.pt.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Hill P, Kessl JJ, Fisher N, Meshnick S, Trumpower BL, Meunier B. Recapitulation in Saccharomyces cerevisiae of cytochrome b mutations conferring resistance to atovaquone in Pneumocystis jiroveci. Antimicrob Agents Chemother. 2003;47:2725–2731. doi: 10.1128/AAC.47.9.2725-2731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessl JJ, Hill P, Lange BB, Meshnick S, Meunier B, Trumpower BL. Molecular basis for atovaquone resistance in Pneumocystis jirovecii modeled in the cytochrome bc1 complex of Saccharomyces cerevisiae. J Biol Chem. 2004;279:2817–2824. doi: 10.1074/jbc.M309984200. [DOI] [PubMed] [Google Scholar]
- 12.McFadden DC, Tomavo S, Berry EA, Boothroyd JC. Characterization of cytochrome b from Toxoplasma gondii and Qo domain mutants as amechanism of atovaquone-resistance. Mol Biochem Parasitol. 2000;108:1–12. doi: 10.1016/s0166-6851(00)00184-5. [DOI] [PubMed] [Google Scholar]
- 13.Hughes LM, Covian R, Gribble GW, Trumpower BL. Probing binding determinants in center P of the cytochrome bc1 complex using novel hydroxynaphthoquinones. Biochim Biophys Acta. 2010;1797:38–43. doi: 10.1016/j.bbabio.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fieser LF, Berliner E, Bondhus FJ, Chang FC, Dauben WG, Ettlinger MG, Fawaz G, Fields M, Fieser M, Heidelberger C, Heyman H, Seligman AM, Vaughan WR, Wilson AG, Wilson E, Wu MI, Leffler MJT, Hamlin KE, Hathaway RJ, Matson EJ, Moore EE, Moore MB, Rapala RT, Zaugg HE. Naphthoquinone antimalarials: general survey. J Amer Chem Soc. 1948;70:3151–3155. doi: 10.1021/ja01190a001. [DOI] [PubMed] [Google Scholar]
- 15.Fieser LF, Chang FC, Dauben WG, Heidelberger C, Heymann H, Seligman AM. Naphthoquinone anti-malarials XVIII. Metabolic oxidation products. J Pharmacol Exptl Therapeut. 1948;94:85–96. [PubMed] [Google Scholar]
- 16.Kessl JJ, Moskalev NV, Gribble GW, Nasr M, Trumpower BL. Parameters determining the relative efficacy of hydroxynaphthoquinone inhibitors of the cytochrome bc1 complex. Biochim Biophys Acta. 2007;1767:319–326. doi: 10.1016/j.bbabio.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessl JJ, Ha KH, Merritt AK, Meshnick SR, Trumpower BL. Molecular basis of Toxoplasma gondii atovaquone resistance modeled in Saccharomyces cerevisiae. Mol Biochem Parasitol. 2006;146:255–258. doi: 10.1016/j.molbiopara.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt ME, Trumpower BL. The petite phenotype resulting from a truncated copy of subunit 5 results from loss of assembly of the cytochrome bc1 complex and can be suppressed by over-expression of subunit 9. J Biol Chem. 1991;266:14958–14963. [PubMed] [Google Scholar]
- 19.Snyder C, Trumpower BL. Mechanism of ubiquinol oxidation by the cytochrome bc1 complex: Pre-steady-state kinetics of cytochrome bc1 complexes containing site-directed mutants of the Rieske iron-sulfur protein. Biochim Biophys Acta. 1998;1365:125–134. doi: 10.1016/s0005-2728(98)00052-8. [DOI] [PubMed] [Google Scholar]
- 20.Yu CA, Yu L, King TE. Preparation and properties of cardiac cytochrome c1. J Biol Chem. 1972;247:1012–1019. [PubMed] [Google Scholar]
- 21.Berden JA, Slater EC. The reaction of antimycin with a cytochrome b preparation active in reconstitution of the respiratory chain. Biochim Biophys Acta. 1970;216:237–249. doi: 10.1016/0005-2728(70)90215-x. [DOI] [PubMed] [Google Scholar]
- 22.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milhous WK, Weatherly NF, Bowdre JH, Desjardins RE. In vitro activities of and mechanisms of resistance to antifol antimalarial drugs. Antimicrob Agents Chemother. 1985;27:525–530. doi: 10.1128/aac.27.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacci JB., Jr Inhibition of liver-stage development assay. Methods Mol Med. 2002;72:517–520. doi: 10.1385/1-59259-271-6:517. [DOI] [PubMed] [Google Scholar]
- 25.Ozaki LS, Gwadz RW, Stable GNG. Simple centrifugation method for rapid separation of sporozoites from mosquitoes. J Parasit. 1984;70:831–833. [PubMed] [Google Scholar]
- 26.Janse CJ, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, Matuschewski K, van Gemert GJ, Sauerwein RW, Waters AP. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006;145(1):60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Caridha D, Yourick D, Cabezas M, Wolf L, Hudson TH, Dow GS. Mefloquine-induced disruption of calcium homeostasis in mammalian cells is similar to that induced by ionomycin. Antimicrob Agents Chemother. 2008;52(2):684–693. doi: 10.1128/AAC.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solmaz SRN, Hunte C. Structure of Complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem. 2008;283:17542–17549. doi: 10.1074/jbc.M710126200. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 30.Smilkstein MJ, Forquer I, Kanazawa A, Kelly JX, Winter RW, Hinrichs DJ, Kramer DM, Riscoe MK. A drug-selected Plasmodium falciparum lacking the need for conventional electron transport. Mol Biochem Parasitol. 2008;159:64–68. doi: 10.1016/j.molbiopara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merbitz-Zahradnik T, Zwicker K, Nett JH, Link TA, Trumpower BL. Elimination of the disulfide bridge in the rieske iron-sulfur protein allows assembly of the [2Fe-2S] cluster into the rieske protein but damages the ubiquinol oxidation site in the cytochrome bc1 complex. Biochemistry. 2003;42:13636–13645. doi: 10.1021/bi035344r. [DOI] [PubMed] [Google Scholar]
- 32.Bowyer JR, Edwards CA, Ohnishi T, Trumpower BL. An analogue of ubiquinone which inhibits respiration by binding to the iron-sulfur protein of the cytochrome bc1 segment of the mitochondrial respiratory chain. J Biol Chem. 1982;257:8321–8330. [PubMed] [Google Scholar]
- 33.Palsdottir H, Gomez Lojero C, Trumpower BL, Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 34.Covian R, Kleinschroth T, Ludwig B, Trumpower BL. Asymmetric binding of stigmatellin to the dimeric Paracoccus denitrificans bc1 complex: evidence for anti-cooperative ubiquinol oxidation and communication between center P ubiquinol oxidation sites. J Biol Chem. 2007;282:22289–22297. doi: 10.1074/jbc.M702132200. [DOI] [PubMed] [Google Scholar]
- 35.Ding MG, Butler CA, Saracco SA, Fox TD, Godard F, di Rago JP, Trumpower BL. Introduction of cytochrome b mutations in Saccharomcyes cerevisiae by a method that allows selection for both functional and non-functional cytochrome b Proteins. Biochim Biophys Acta. 2008;1777:1147–1156. doi: 10.1016/j.bbabio.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakato H, Vivancos R, Hunter PR. A systematic review and meta-analysis of the effectiveness and safety of atovaquone–proguanil (Malarone) for chemoprophylaxis against malaria. J Antimicrob Chemo. 2007;60:929–936. doi: 10.1093/jac/dkm337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.