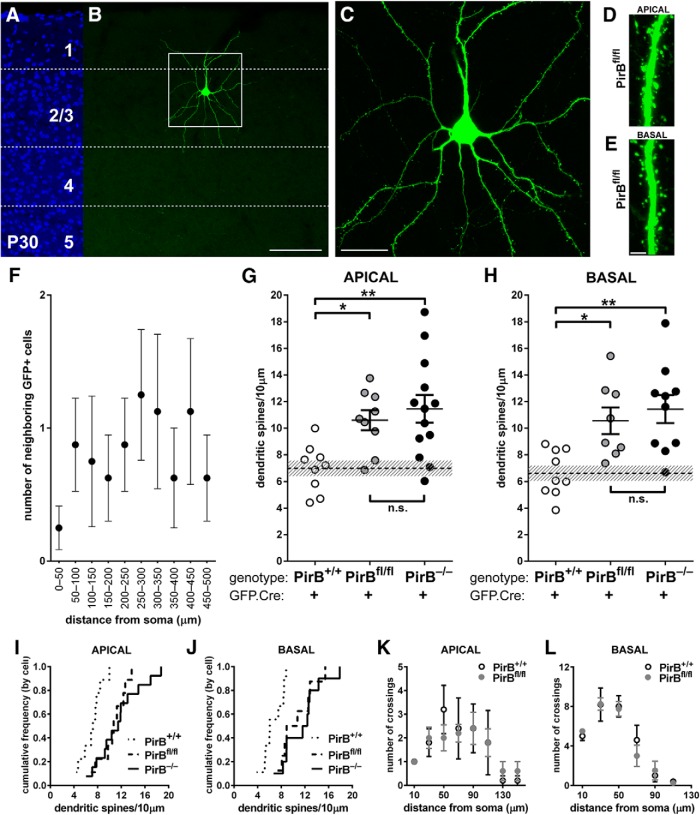
Figure 2.
Dendritic spine density at P30 is elevated on isolated PirB−/− neurons in layer 2/3 after sparse excision of PirB at E15.5. A, B, Fluorescent micrographs at P30 of nuclear counterstain (DAPI; A) and an isolated L2/3 pyramidal neuron electroporated with GFP.Cre (B) from a visual cortex section from a P30 PirBfl/fl mouse. C, High-magnification maximum intensity projection of the boxed area in B. D, E, Zoomed-in high-magnification micrographs of portions of apical (D) and basal (E) dendrites showing dendritic spines in PirBfl/fl mice. F, Number of neighboring GFP+;Cre+ cells as a function of distance from a neuron of interest in PirBfl/fl tissue (n = 8 cells, 7 mice). The graph shows that, on average, there was only one GFP+;Cre+ cell in every 50 μm increment analyzed. G, Spine density on apical dendrites is greater in GFP+;Cre+ neurons from PirBfl/fl mice than PirB+/+ mice (PirB+/+: 7.0 ± 0.6 dendritic spines/10 μm of dendritic length, n = 9 cells, 5 mice; same data as in Fig. 1I; PirBfl/fl, GFP+;Cre+: 10.6 ± 0.8, n = 9 cells, 7 mice; PirB−/−, GFP+;Cre+: 11.5 ± 1.0, n = 13 cells, 5 mice; same data as in Fig. 1I; PirB+/+ vs PirBfl/fl, p = 0.041a; PirBfl/fl vs PirB−/−, p = 1.000a, one-way ANOVA with post hoc Bonferroni’s multiple comparisons). H, Basal dendritic spine density is greater in GFP+; Cre+ neurons from PirBfl/fl mice than PirB+/+ mice (PirB+/+: 6.6 ± 0.6 dendritic spines/10 μm of dendritic length, n = 9 cells, 5 mice; data from Fig. 1J; PirBfl/fl, GFP+;Cre+: 10.6 ± 1.0, n = 8 cells, 7 mice; PirB−/−, GFP+;Cre+: 11.4 ± 1.1, n = 10 cells, 5 mice; data from Fig. 1J; PirB+/+ vs PirBfl/fl p = 0.02b, PirBfl/fl vs PirB−/− p = 1.000b, One-way ANOVA with post hoc Bonferroni's multiple comparisons). I, Cumulative histogram (by cell) of data presented in G. J, Cumulative histogram (by cell) of data presented in H. K, L, Sholl analysis reveals no significant changes in apical (K) or basal (L) dendritic branching between GFP+;Cre+ L2/3 neurons from PirB+/+ and PirBfl/fl mice (K: PirB+/+, n = 5 cells, 3 mice; data from Fig. 1K; PirBfl/fl, n = 5 cells, 3 mice, p = 0.8864e; L: PirB+/+, n = 5 cells, 3 mice; data from Fig. 1L; PirBfl/fl, n = 4 cells, 3 mice, p = 0.7799f, two-way ANOVA with repeated measures). *p < 0.05; **p < 0.01. Scale bars: A, B, 100 μm; C, 25 μm; D, E, 3 μm.