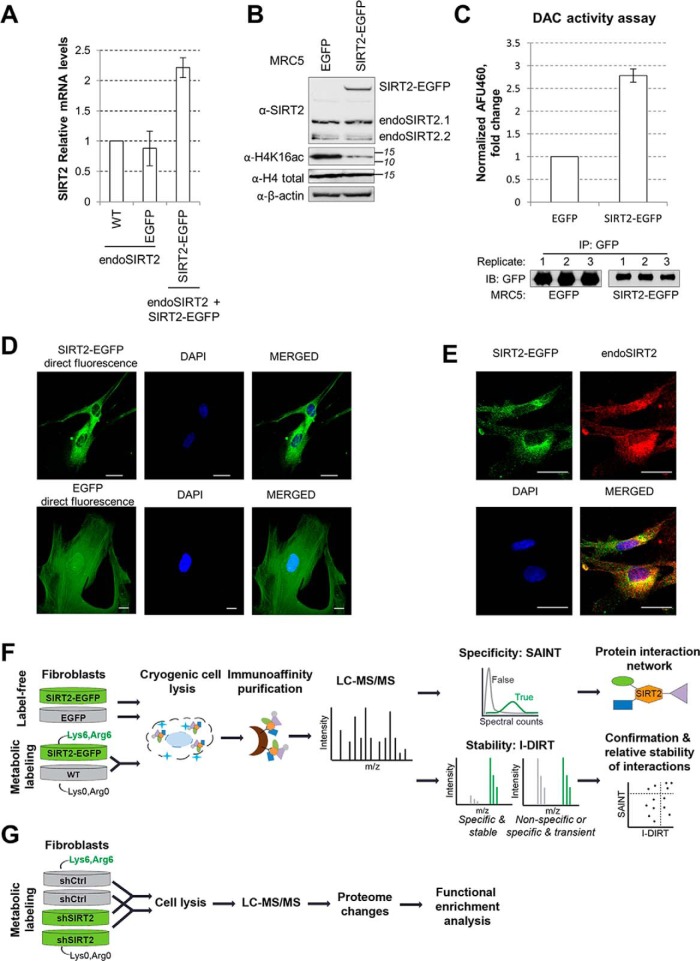
Fig. 1.
Experimental design for characterizing SIRT2 interactions in human fibroblasts. A, SIRT2-EGFP is localized to the cytoplasm. MRC5 cells stably expressing SIRT2-EGFP and EGFP were imaged by direct fluorescence microscopy, using DAPI as nuclear marker (blue). Bar size = 25 μm, 63Xmag. B, SIRT2-EGFP localizes similarly to endogenous SIRT2. MRC5 SIRT2-EGFP were imaged by immunofluorescence microscopy using anti-GFP (green) and anti-SIRT2 (red) antibodies, and DAPI nuclear stain (blue). Bar size = 25 μm, 63Xmag. Max Projection. C, SIRT2 mRNA levels in wt, EGFP, and SIRT2-EGFP MRC5 cells. SIRT2 levels were normalized to actin levels across all samples, n = 2. D, SIRT2 protein levels in EGFP and SIRT2-EGFP MRC5 cells. SIRT2-EGFP and endogenous SIRT2 were detected in MRC5 EGFP and SIRT2-EGFP whole-cell lysates using anti-SIRT2 antibody. Histone H4K16ac is a known SIRT2 substrate. Total histone H4 and actin are loading controls. E, SIRT2-EGFP construct is catalytically active. Deacetylase activity of SIRT2-EGFP and EGFP alone was measured upon IP from MRC5 cells using in vitro fluorometric assay. Western blotting using anti-GFP antibody shows equal isolation levels for each protein in the replicates (n = 3). F, Workflow using label-free and metabolic labeling approaches for identifying SIRT2 interactions, their specificity and relative stability. G, Workflow using metabolic labeling for determining changes in the cellular proteome upon shRNA-mediated knockdown of SIRT2.