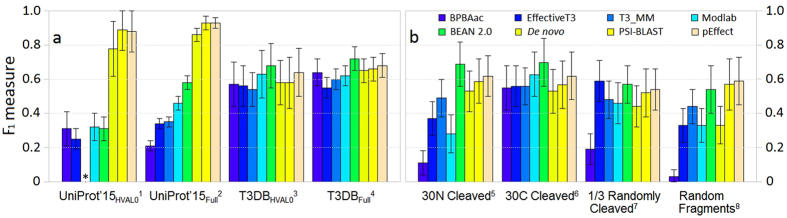
Figure 1. Method performance comparison on independent test sets and protein fragments.
Performance (Supplementary S1 Text: F1 measure, Eqn. 3; ‘ ± ’ standard error, Eqn. 4) was measured for BPBAac, EffectiveT3, T3_MM, Modlab and BEAN 2.0 methods (Supplementary S2 Text). We also computed F1 for de novo (SVM-based) predictions alone, PSI-BLAST homology-based look up alone, and pEffect: a combination of PSI-BLAST (if a hit is available) and de novo (otherwise). Panel (a) shows performance on evaluation data sets (Methods) including (1)UniProt’15HVAL0 (51 effectors and 691 non-effector bacterial and eukaryotic proteins, added to UniProt after 2014_02 release, sequence homology reduced at HVAL < 0), (2)UniProt’15Full (498 effectors and 1,509 non-effector bacterial and eukaryotic proteins added to UniProt after 2014_08 release, NOT homology reduced), (3)T3DBHVAL0 (66 effectors and 128 non-effector bacterial proteins from T3DB database, sequence homology reduced at HVAL < 0), and (4)T3DBFull (218 effectors and 831 non-effector bacterial proteins from T3DB database, NOT homology reduced). Note: T3_MM was not able to produce results for the UniProt’15HVAL0 set during manuscript preparation. Panel (b) shows performance on fragments produced from (3)T3DBHVAL0 (Methods) including (5)approach i: 30 N-terminal amino acids cleaved off, (6)ii: 30 C-terminal amino acids cleaved off, (7)iii: Randomly selected two thirds of the protein sequence, and (8)iv: Randomly selected sequence fragments of typical translated read length (average 110 amino acids, Supplementary Fig. S1).