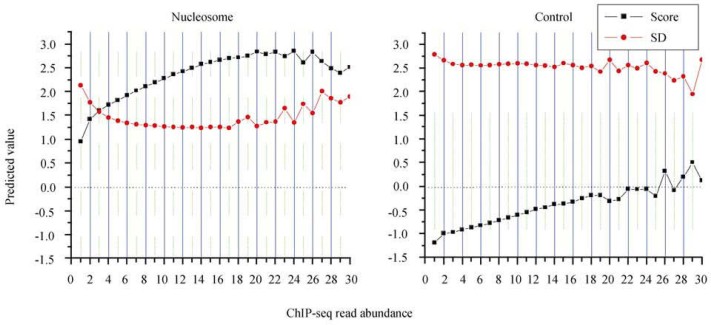
Figure 7.
Prediction of nucleosome positioning based on the HMM algorithm. We first use nucleosome-protected sequences from the fifth chromosome (the largest, ~20 Mb) as a training set to obtain frequencies of state transition and emission (see Figure 6B). We then apply the HMM algorithm on the nucleosome-protected sequences from the rest five chromosomes. Each nucleosome-protected sequence is assigned a prediction score based on the HMM algorithm. A score above zero indicates that the corresponding sequence has a tendency to be nucleosome-associated, whereas a minus score suggests nucleosome-free. We group the sequences based on ChIP-seq read abundance and calculate the average prediction score as well as its standard deviation (SD). Note that average prediction scores for the nucleosome-protected sequence groups are all above zero, as opposed to minus scores for the control. Prediction scores are proportional to read abundance.