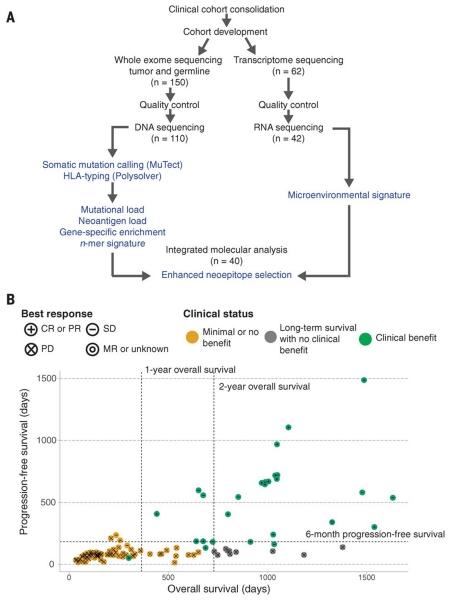
Fig. 1. Study design and clinical stratification.
(A) Patients (n = 150) were identified for whole-exome sequencing of tumor and germline DNA. To be included in the original clinical cohort, patients had to have received ipilimumab monotherapy for metastatic cutaneous melanoma, have pretreatment germline and tumor samples available for sequencing, and have had overall survival for >14 days after initiation of ipilimumab therapy. Of these patients, 110 were eventually included in analysis after exclusions due to inadequate postsequencing quality control (n = 40) (18). Manual review of raw sequencing data was performed to exclude samples with evidence suggesting low purity, high contamination by ContEst (33), or discordant copy number quality control. Of the patients, 62, including 2 who failed DNA quality-control, had FFPE tumor samples available for transcriptome sequencing. After manual review for quality control following RNA sequencing, 42 samples were also analyzed for tumor microenvironment signatures, and 40 with matched WES were analyzed for neoantigen expression (14). (B) Patients were stratified into response groups based on RECIST criteria (21) (CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; MR, mixed response); duration of overall survival (OS); and duration of progression-free survival (PFS). All two-way comparisons were done comparing patients who achieved clinical benefit with ipilimumab (CR or PR by RECIST criteria or OS >1 year with SD by RECIST criteria) (n = 27) to those with minimal or no benefit from ipilimumab (PD by RECIST criteria or OS <1 year with SD by RECIST criteria) (n = 73). An additional cohort of patients who achieved long-term survival (OS >2 years) after ipilimumab treatment with early tumor progression (PFS <6 months) were considered separately (n = 10).