Abstract
Enzymes catalyze various biochemical functions with high efficiency and specificity. In vitro design of the enzyme leads to novel bioactivity in this natural biomolecule that give answers of some vital questions like crucial residues in binding with substrate, molecular evolution, cofactor specificity etc. Enzyme engineering technology involves directed evolution, rational designing, semi-rational designing, and structure-based designing using chemical modifications. Similarly, combined computational and in vitro evolution approaches together help in artificial designing of novel bioactivity in the natural enzyme. DNA shuffling, error prone PCR and staggered extension process are used to artificially redesign active site of enzyme, which can alter its efficiency and specificity. Modifications of the enzyme can lead to the discovery of new path of molecular evolution, designing of efficient enzymes, locating active sites and crucial residues, shift in substrate, and cofactor specificity. The methods and thermodynamics of in vitro designing of the enzyme are also discussed. Similarly, engineered thermophilic and psychrophilic enzymes attain substrate specificity and activity of mesophilic enzymes that may also be beneficial for industry and therapeutics.
Keywords: in vitro design, strategic manipulation, molecular evolution, novel bioactivity of natural enzyme, thermodynamics of in vitro design
Introduction
Enzymes are the biocatalysts that have specificity, efficiency, and mode of regulation. They are also used for varied industrial processes especially in medical research such as de novo drug discovery and its improvement (increase the yield of antibiotics, hormones, etc.), crop improvement, biofuel industry, food industry, etc. Enzymes used in biotechnological process have to be very efficient in their catalytic activity because in vivo and in vitro conditions are very different. Most of the enzymes are derived from mesophilic organisms; hence they are most active in moderate conditions. Therefore, in order to improve the catalytic performance of the industrial enzymes and withstand harsh conditions, protein engineering plays a major role, and resulting in the reduction of cost and time of processing in the respective industries. Changing cofactor specificity, discovering path of molecular evolution, designing more powerful and efficient enzymes, locating active sites and crucial residues, shifting the substrate specificity toward new substrate, improving binding affinity, changing in pH tolerance and inhibitor tolerance, improving solubility and resistance to extreme conditions, are some of the modifications that can be achieved by in vitro enzyme designing. Enzymes can be altered by redesigning the active site that can alter its efficiency (kcat/km) and specificity (km). Altering the ligand binding sites, incorporation of new functional groups into the active site, DNA shuffling, site-saturation, and site-directed mutagenesis are some of the approaches that modify the active site of an enzyme. Thermo-stabilizing traits such as high number of hydrogen bonds, introduction of ion pairs for linking of N and C-terminals, addition of proline residues in loops are among many modifications that can be achieved at genetic level through mutagenesis after complete analysis of sequence and structure of homologous mesophilic and thermophilic enzymes. Cryo-stabilizing characteristics such as clustering of glycine residues which provide local flexibility in the region, reduced frequency of proline residues in loops leading to enhanced chain flexibility between secondary structures, a reduction in arginine residues which generally participate in formation of multiple salt bridges and H-bonds, and a lowered number of ion pairs and aromatic interactions, can also be introduced by mutagenesis. These characteristics of thermophilic and cryophilic enzyme can be added in the mesophilic enzyme.
Theodosius Dobzhansky stated that “Nothing in biology makes sense except in the light of evolution” (Dobzhansky, 1973). Evolution is the change in heritable phenotypic trait of biological population over successive generations. The amazing similarity in gene sequences and metabolic pathways across the phyla says that modern organisms are derived from common progenitor by series of small changes, i.e., mutations. Homology means two proteins are descended from a common ancestor while sequence homology means amino acids at equivalent positions are similar because they are descended from a common ancestor (Hall and Barlow, 2004). The gradual accumulation of mutations over long period of time results in the formation of new species. Natural in vivo design of the bioactivity leads to the molecular evolution. Molecular evolution is a fundamental biological process and its understanding would facilitate the engineering of enzymes with new specificities. During the course of evolution, it is possible that inefficient enzymes convert to efficient enzymes or vice-versa (Geddie and Matsumura, 2004). The in vitro design of the activity can be done using various mutagenesis approaches. Thus, enzyme engineering using in vitro approaches may contribute to elucidate structural basis of the functional properties, and the course of the natural molecular evolution. In the present review, we have explained the methods for In vitro mutagenesis and their significances in the enzyme substrate replacement, cofactor replacement, and change in enzyme catalysis.
Methods for in vitro design or artificial mutation
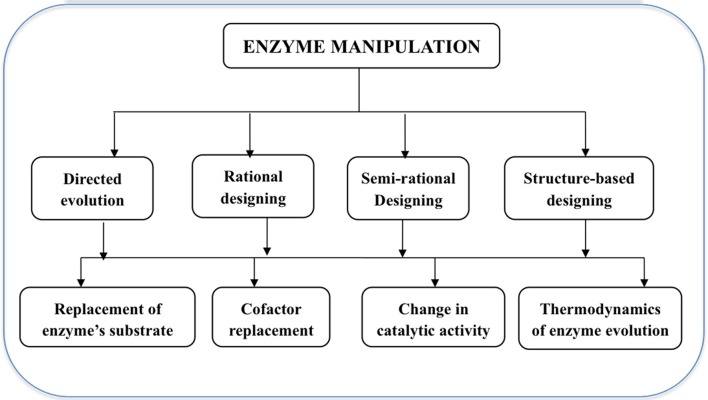
Protein engineering technology (Figure 1) involves directed evolution, rational designing, semi-rational design, structure-based design using chemical modification, and truncated and fusion methods (Li and Cirino, 2014; Kaushik et al., 2016). In directed evolution, natural evolution is mimicked in the lab and based on developing molecular diversity by random mutagenesis and in vitro recombination followed by performing high-throughput screening for improvements in desired phenotype. Similarly, rational protein design is used for those enzymes whose three-dimensional structures have already been determined and site-directed mutagenesis is used to introduce specific and deliberate mutations on the enzyme sites. Both directed evolution and rational protein design have certain limitations, hence a combined approach known as semi-rational design, takes advantage of already available information of structure and function of the desired protein and designs a smaller but higher quality library. Molecular dynamics and QM studies also compliment such studies as they facilitate the understanding about the impact of every single amino acid on the structure and function of the protein. This computer-guided semi-rational designing of proteins further helps in shaping the needs of a good biocatalyst with a larger substrate range, selectivity, specificity, and stability without harming their catalytic efficiency (Kaushik et al., 2016).
Figure 1.
Summary of enzyme engineering methods and their significances.
Recent studies have shown that combined computational and in vitro evolution approaches can be used for the study of evolution of novel bioactivity in the natural enzymes. In vitro study can be achieved by construction of large libraries of protein/enzyme mutants, enzyme with certain substrate binding characteristics are selected, and the sequences of the selected enzyme are determined. The main advantage of this method is that they require minimal knowledge of enzyme substrate interaction under study. This method does not provide much information about the nature of intermolecular contacts, which often hampers an understanding of the obtained results. Computational protein design is parallel approach that is based on our understanding of the biophysical forces that govern enzyme-substrate binding but it require a detailed knowledge of the structure and function of the enzyme substrate under study (Rosenfeld et al., 2016).
Other approach using novel scaffolding and compartmentalization techniques are also used to improve performance of enzyme (Li and Cirino, 2014). Consensus design is another approach that can be used to artificial designing of enzyme, which is based on the hypothesis that at a given position of enzyme, the respective consensus amino acid contributes more than average to the stability of the enzyme than non-conserved amino acids. Consensus design, like ancestral sequence reconstruction, utilizes evolutionary history; all sequences are aligned, and most frequently observed amino acid is identified at each position in the alignment (Porebski and Buckle, 2016). To achieve the molecular evolution of a protein, a consensus library for directed evolution experiments could be made, which would contain information about site-directed mutations of a particular protein. This mimics the natural Darwininan evolution process, which involves repeated cycles of mutation, followed by selection that renders a strong algorithm to create diversity as exhibited by the plethora of life. The process can be performed in vitro by introducing random mutations on template strand, followed by screening and selection of desired mutant clones. The whole procedure is repeated until desired mutation is achieved. Selection or screening of a consensus library should reveal correlative or co-varying substitutions that change parallel in order to alter protein function (Flores and Ellington, 2005). Redesigning of enzymes does not only require change in the residues that bind to substrates and cofactor but also requires residues that are far from active site but important to maintain catalytic residues in prerequisite orientations. Electrostatic charge distribution throughout a whole enzyme molecule may also facilitate substrate binding by electrostatically guiding the substrate into the active site (Oue et al., 1999). The mutagenized gene library is made by various methods such as DNA shuffling, error prone PCR (ePCR) and staggered extension process (StEP). However, the efficiency rate of directed evolution is low and less yield of the final PCR product acts as a drawback for the downstream processing. This can be avoided using ExoSAP-IT PCR Product Cleanup (Affymetrix), that serves as an alternative to beads based PCR (Goh et al., 2012).
ePCR incorporates error in the final product during DNA amplification. Errors are introduced by altering the parameters of PCR such as use of high concentrations of magnesium or manganese, DNA polymerase not capable of proofreading, increase in the number of amplification cycles and/or biased concentration of dNTPs. DNA shuffling is one of the most important directed evolution techniques which has increased the catalytic efficiency of various industrial enzymes namely β- 1,3- 1,4-glucanase (Yan et al., 2006), glucose dehydogenase (Gruber et al., 1998) etc. This technique involves formation of various chimeras by joining different homologous genes via self-priming PCR and ultimately creating a library. More than 80% similarities should be there in homologous parent genes (Brakmann and Schwienhorst, 2004). StEP is also a commonly used homology-dependent process for performing in vitro DNA recombination. For the purpose of efficient recombination the parental genes must share more than 85% sequence identity (Volkov and Arnold, 2000).
Recently, mutagenesis methods have improved tremendously in comparison to earlier laborious primer extension methods that require templates (to be mutated) in single stranded form as well as stringent screening procedures for selection of mutants. Advance methods such as Exsite and GeneTailor uses double stranded template DNA and PCR to mutate the targets followed by easy selection of mutants. Alanine scan is a standard method to analyze the active site mechanism and identify the role of important residues, where the concerned residues are mutated with alanine (Dembowski and Kantrowitz, 1994). Hence with the types of technologies available today and with the widely accessible information (web based databases), engineering procedures have become routine and are continuously introducing new enzymes and proteins into market with enhanced traits or novel properties for therapeutic, pharmaceutical and industrial applications. In vitro mutagenesis results from the three type of changes i.e., enzyme substrate replacement, cofactor replacement, and change in enzyme catalysis. We have critically analyzes each of these in detail by taking examples of different enzymes. Various enzymes and their mutants formed by site-directed mutagenesis along with the mutation sites are listed in Table 1.
Table 1.
Enzymes and their mutants formed by site directed mutagenesis.
| S. no. | Source | Wild type enzyme | Substrate specificity/cofactor specificity | Mutant enzyme | New substrate specificity/cofactor specificity | Mutations at specific sites | References |
|---|---|---|---|---|---|---|---|
| 1. | Trichomonas vaginalis | Lactate dehydrogenase | Lactate etc. | Malate dehydrogenase | Malate etc. | Leu91Arg | Wu et al., 1999 |
| 2. | Citrobacter freundii | Tyrosine phenol lyase | β- elimination of tyrosine | Dicarboxylic amino acid β-lyase enzyme | Reversible transfer of an amino group from dicarboxylic amino acids to oxo acids | Arg100Thr, Val283Arg | Mouratou et al., 1999 |
| 3. | Citrobacter freundii | Tyrosine phenol lyase | β- elimination of tyrosine | Tyrosine phenol lyase | Decreased affinity for β- elimination of tyrosine | Asn185Ala, Tyr71Phe, Thr124Asp, Phe448His | Chen H. Y. et al., 1995; Barbolina et al., 2000; Demidkina et al., 2002 |
| 4. | Lactobacillus pentosus | NAD-dependent D-lactate dehydrogenase | Pyruvate | D-2hydroxyisocaproate dehydrogenase | Larger aliphatic or aromatic 2-ketoacid substrates | Tyr52Leu | Tokuda et al., 2003 |
| 5. | E. coli | Choline acetyl transferase | Acetyl group acceptor is choline | Carnitine acetyl transferase | Specificity More for carnitine and less for choline | Val459Thr, Asp460Glu, Asn461Thr Asn514Arg | Reznik et al., 1998 |
| 6. | E. coli | Aspartate amino- transferase | reversible transfer of the amino group of aspartate or glutamate to the cognate oxo acids | L-aspartate-β decarboxylase | β-caboxylase activity | Tyr225Arg, Arg292Lys Arg386Ala | Graber et al., 1999 |
| 7. | Bacillus stearothermophilus | Lactate dehydrogenase | specific for cofactor NAD+ | Same | Utilized NADP+ far better | Phe16Gln, Cys81Ser Asn85Arg | Flores and Ellington, 2005 |
| 8. | E. coli | β-Glucuronidase | β- galactosidase activity | Same | Increased in β galactosidase activity | Thr509Ala, Ser557Pro, Asn566Ser Lys568Gln | Geddie and Matsumura, 2004 |
| 9. | E. coli | Malate dehydrogenase | Inter-conversion of malate & oxaloacetate | Same | Catalytic activity is decreased & specificity for other substrate increased | Arg153Cys | Wright and Viola, 2001 |
| 10. | E. coli | Aspartate amino transferases | Aspartate | Same | Five-fold increase in activity | Asn34Asp, Ile37Met, Ser139Gly, Asn142Thr, Asn297Ser, Val387Leu | Yano et al., 1998 |
| 11. | E. coli | Aspartate amino transferases | Aspartate | same | Valine | 17 mutations | Oue et al., 1999 |
| 12. | E. coli | Streptavidin | Biotin | Same | Biotin analogs e.g., Iminobiotin | Asn23Ala Ser27Asp | Reznik et al., 1998 |
| 13. | Klebsiella pneumoniae | 1,2 propanediol oxidoreductase | Coenzyme-NADH | Same | Coenzyme-NADH and NADPH | Asp41Ala, Asp41Gly | Ma et al., 2010 |
| 14. | Bacillus lentus | Subtilisins | Proteolytic activity | Same | Increased proteolytic activity | Lys27Arg/ Asn87Ser/ Val104Tyr/ Asn123Ser /Thr274Ala Asn76Asp/ Asn87Ser/ Ser103Ala/ Val104Ile | Graycar et al., 1999 |
| 15. | Streptomyces spp. | Cholestrol oxidase | Pregnenolone, cholesterol | Same | Increased catalytic activity | Ser379Thr | Toyama et al., 2002 |
| 16. | E. coli | Isocitrate dehydrogenase | Coenzyme-NADP | Same | Coenzyme-NAD | 6 mutations | Chen R. et al., 1995 |
| 17. | Peptostreptococcus asaccharolyticus | Glutamate dehydrogenase | Coenzyme-NADH | Same | Coenzyme-NADPH | Glu243Lys/Glu243Asp | Carrigan and Engel, 2007 |
| 18. | Candida boidinii | Formate dehydrogenase | Coenzyme-NAD+ | Same | Coenzyme-NADP+ | Asp195Gln/Tyr196His | Andreadeli et al., 2008 |
| 19. | E. coli | Malate dehydrogenase | Oxaloacetate | Phenylactate dehydrogenase | Phenyl pyruvate | Arg81Cys | Wright et al., 2000 |
| 20. | Streptomyces coelicolor | Malate dehydrogenase | NADP | same | NADPH | Glu42Gly, Ile43Ser, Pro45Arg, Ala46Ser | Ge et al., 2014 |
| 21. | Pseudomonas N176 | Glutaryl-7-ACA acylase | CephC | CephalosporinCacylases (CA) | CephC | His57Ser His70Ser, Leu154Tyr | Conti et al., 2014 |
| 22. | Candida tenuis | Xylose reductase | NADPH | Same | NADH | Lys274Arg–Asn276Asp | Petschacher et al., 2005 |
| 23. | Thermus thermophilus | Lactate dehydrogenase | NADH | Same | NADPH | 7 amino acid residues | Tomita et al., 2006b |
| 24. | Pichia stipitis | Xylose reductase | NADPH | Same | NADH | Lys21Ala/ Asn272Asp | Zeng et al., 2009 |
| 25. | Clostridium symbiosum | Glutamate dehydrogenase | NADH | Same | NADPH | Asp263Lys, Phe238Ser, Pro262Ser | Griffin and Engel, 2011 |
| 26. | Thermus flavus | Malate dehydrogenase | NADH | Same | NADPH | 7 amino acids residues | Nishiyama et al., 1993; Tomita et al., 2006a |
| 27. | E. coli | Lactaldehyde dehydrogenase | NADH | Same | NADPH | Phe180Thr | Rodríguez-Zavala, 2008 |
| 28. | Candida magnoliae | Carbonyl reductase | NADPH | Same | NADH | 8 amino acid residues | Morikawa et al., 2005 |
| 29. | Neurospora crassa | Nitrate reductase | NADPH | Same | NADH | Ser920Asp Arg932 Asp | Shiraishi et al., 1998 |
| 30. | Lactobacillus bulgaricus | Lactate dehydrogenase | NADH | Same | NADPH | Asp175Ala | Bernard et al., 1995 |
| 31. | Glomerella cingulata | Cutinase | Hydrolysis of esters and triglycerides | Same | Catalytically inactive | His204Asn | Nyon et al., 2009 |
| 32. | Pseudomonas aeruginosa LST- 03 | Lipase | Ester synthesis and inter-esterification reaction and lipid hydrolysis | Same | Increased organic solvent stability | Ser155Leu Gly157Arg, Ser164Lys, Ser194Arg, Asp209Asn | Kawata and Ogino, 2010 |
| 33. | Geobacillus kaustophilus HTA426 | Lactonase | 3-oxo-N-dodecanoyl-L-homoserine lactone | Same | 72-fold increase in the catalytic efficiency | Glu101Asn/ Arg230Ile | Chow et al., 2010 |
| 3-oxo-N-dodecanoyl-L-homoserine lactone | Same | N-butyryl-l-homoserine lactone | Asp266Asn | Chow et al., 2010 | |||
| 34. | alkalophilic Bacillus sp. | Cyclodextrin glucano transferase | produce cyclodextrins from starch | Same | higher product yields | His233Tyr | Leemhuis et al., 2010 |
| 35. | E. coli | Lactaldehyde reductase | NADH | Same | 3.6-fold increase in kcat | Met185Cys | Cahn et al., 2016 |
| 36. | Saccharomyces cerevisae | Cinnamyl alcohol dehydrogenase | NAD(P)H | Same | 82-fold increase in activity | Gln110Val | Cahn et al., 2016 |
| 37. | Actinoplanes utahensis | Cephalosporin acyclase | Cephalosporin | Same | Aculeacin A | Phe177(β)Gly/Met145(α)Ala and Phe177(β)Gly/Met145(α)Ala/Tyr149(α)Val | Isogai and Nakayama, 2016 |
Significance of in vitro manipulation
Replacement of natural substrate with a novel substrate
The alteration in the substrate specificity is seen mainly in the dehydrogenases. The reason might be the utilization of similar cofactor i.e., NAD by all dehydrogenases. Malate dehydrogenase (MDH) from Escherichia coli is highly specific for its keto acid substrate. The replacement of one of the binding arginine residues (81st position) with unnatural aryl and alkyl amino acid analogs, result into enhancement of the specificity of MDH for phenyl pyruvate while diminished for oxaloacetate (physiological substrate; Wright et al., 2000). This is performed by site-specific modulation that incorporates systemic structural information at the target site. Similarly, some experiments also revealed that three arginines (80th, 87th, and 153th) of malate dehydrogenase are responsible for selectivity of the substrate. Site-directed mutagenesis at Arg153Cys was done which decreased its specificity for malate and oxaloacetate. This substitution changed the conformation of the active site and thus the specificity for original substrate decreased. Substitutions of functional group like acetamide etc. changed the substrate specificity to pyruvate, alpha-ketoglutarate etc. (Bell et al., 2001; Wright and Viola, 2001).
Some other studies reported that a single amino acid mutation (Tyr52Leu) was done in NAD-dependent D-lactate dehydrogenase of Lactobacillus pentosus, resulted in the increased affinity of enzyme toward larger aliphatic or aromatic 2-ketoacid substrates by four-fold as well as decreased affinity toward pyruvate (which is its actual substrate) by about 30-fold and thus converting the enzyme into D-2-hydroxyisocaproate dehydrogenase (an active enzyme; Tokuda et al., 2003).
Likewise, many attempts have been made to redesign the active sites of enzymes such as lactate dehydrogenase (LDH) present in Trichomonas vaginalis (Tv). It resembles malate dehydrogenase (cytosolic) more closely than LDH of other species. The Arg at 91st position of MDH was replaced by Leu in TvLDH. The change Leu91Arg was done by site directed mutagenesis that converted TvLDH to MDH. The reverse change i.e., Arg91Leu does not yield a measurable activity of TvLDH. The result was verified by molecular modeling and in vitro functional characterization. This conversion showed that MDH-LDH barrier has been broken in at least one direction. This also highlighted the role of some important residue for selection of catalysis of a substrate. Phylogenetic reconstructions showed that members of the MDH and LDH families form separate monophyletic lineages probably arose from early gene duplication (Wu et al., 1999). Similarly, studies suggest that evolution of (Cryptosporidium parvum) lactate dehydrogenase from malate dehydrogenase arose by gene duplication. Ancestral sequence reconstruction technique is useful to study these evolutionary paths (Madern et al., 2004).
Another enzyme, aspartate aminotransferases are the most extensively studied vitamin B-6 containing enzyme. These are homodimeric enzymes that catalyze amino group transfer between acidic amino acids, aspartate and glutamate, and their corresponding 2-oxo acids. The triple mutation at its substrate-binding site leads to increased rate of L-aspartate β-decarboxylase activity to transaminase activity by 25 million-fold. In order to elucidate both the structural basis of the functional properties and course of the molecular evolution, molecular dynamics, and crystal studies are performed (Graber et al., 1999). Similarly, directed molecular evolution was done by mutating 17 amino acids residues of enzyme aspartate aminotransferases in E. coli. This had resulted in six-fold increase in activity for non-native substrate i.e., valine. The remodeled enzyme had altered subunit interface, and a shift in the enzyme domain. These results clearly demonstrate the importance of the cumulative effects of residues remote from the active site and present a new line of approach to redesign enzymes (Oue et al., 1999).
Molecular dynamics and homology data also identified the critical residues of tyrosine phenol lyase (TPL), an enzyme involved in the β-elimination of tyrosine. Double mutations at R100T and V283R are responsible for switching the substrate specificity of TPL from L-tyrosine to dicarboxylic amino acids in Citrobacter freundii. These mutations increased its β-elimination activity toward dicarboxylic acids by four-fold without destroying its catalytic apparatus. Thus, this experiment gave a novel enzyme dicarboxylic amino acid β-lyase, which is not found in nature. From this study, it can be concluded that these enzymes originated from region-specific catalysts, which first specialized for reaction specificity and then for substrate specificity (Mouratou et al., 1999).
Site directed mutagenesis has been routinely used to redesign enzymes, but the achievement in this technique relies basically on the availability of 3-D structure protein maps. Others techniques that are based on cross-referencing substrate structure and comparisons with protein alignment data, were also used. Choline acetyltransferases reversibly transfer acetyl group between acetyl-coA and choline. After site-directed mutagenesis the mutant enzyme have more affinity for carnitine rather than choline. Thus, novel enzyme called carnitine/choline acetyltransferase was obtained (Cronin, 1998). Likewise, catalytic efficiency of cholesterol oxidase from Streptomyces and Brevibacterium, was also studied by site directed mutagenesis and found that efficiency was increased in the mutant S379T toward cholesterol and pregnenolone. These findings provide ideas for designing more efficient enzyme which can be clinically useful for cholesterol estimation (Toyama et al., 2002).
A well-known industrial biocatalyst Glutaryl acylase (GA), belongs to the N-terminal hydrolase class of hydrolytic enzymes and show wide substrate specificity. CA (cephalosporin C acylase) and GAs are members of the glutaryl acylase family that specifically use CephC as their substrate. GA undergoes a two-step reaction to yield cephalosporin and CA undergoes only one step reaction to yield cephalosporin. CAs isolated from Pseudomonas strains has very low activity for biotechnological use. By using protein engineering approach based on combined use of error-prone PCR mutagenesis, molecular modeling, site saturation and site-directed mutagenesis, the glutaryl acylase from Pseudomonas N176 has been mutated (A215αY-H57βS-H70βS and H57βS-H70βS-L154βY). These enzyme variants have 100-fold increase in specificity constant for CephC (Conti et al., 2014).
In addition to the alteration in the enzymes action, artificial mutation can also alter the proteins for non-catalytic function. Streptavidin and biotin is the model system to study structure and thermodynamic properties of high affinity receptor complexes with its ligand. Biotin binding sites were redesigned in such a way that the major structural motifs of the natural streptavidin-biotin complex are preserved. This was done by changing amino acid residues deep in the biotin-binding pocket to reduce the biotin-binding affinity considerably without disturbing the binding affinities for biotin analogs. Thus, a mutant of streptavidin was made which has greater binding specificity to 2-iminobiotin than to its natural ligand biotin. This approach could also be useful for in vivo targeting applications (Reznik et al., 1998).
Alteration in coenzyme specificity of enzyme
Cofactor specificity can also be altered by using techniques like site-directed mutagenesis, DNA shuffling etc. The enzyme lactate dehydrogenase has a specific cofactor NAD+. Using DNA shuffling method, many mutants were made by substitution at cofactor binding sites. A consensus library was made to study cofactor specificity of Bacillus stearothermophilus lactate dehydrogenase. A triple mutant was selected which had decreased specificity for NAD+ and increased for NADP+. Thus, an enzyme with change in cofactor was made without decrease in its catalytic activity (Flores and Ellington, 2005). Similarly, the NAD-dependent D-lactate dehydrogenase from Lactobacillus bulgaricus had also been modified to display 40-fold shift of coenzyme specificity from NADH to NADPH (Bernard et al., 1995).
Similar experiments were also done in different organisms to change the cofactor specificity. Malate dehydrogenase (MDH) of Streptomyces coelicolor (ScMDH) was altered by site-directed mutations in the Rossman fold region to change its cofactor specificity from NADH to NADPH. The coenzyme specificity (Kcat/Km) of the mutant enzyme was examined and found to be shifted 2231.3-fold toward NADPH (Ge et al., 2014). Similarly, in Thermus thermophilus, Thermus flavus, and Thermococcus kodakarensis replacement of specific amino acids in Rossman fold of NADH dependent lactate dehydrogenase changed its cofactor from NADH to NADPH (Nishiyama et al., 1993; Tomita et al., 2006a; Morimoto et al., 2014).
Xylose reductase (NADPH) and xylitol dehydrogenase (NADH) of Pichia stipiti are involved in the catabolism of xylose and for ethanol production from it. As the cofactors of both these enzymes are different, redox imbalance occurs. To solve this problem, site directed mutagenesis was done at Lys21 in xylose reductase. Both K21A and K21A/N272D mutants preferred NADH over NADPH. Thus, a complete reversal of coenzyme specificity toward NADH and improved catalytic efficiency were achieved for this enzyme (Zeng et al., 2009). In Candida tenuis, similar experiments (single mutant K274R, K274M, K274G, S275A, N276D, R280H, and double mutant K274R-N276D) were done to change the cofactor specificity of xylose reductase (Petschacher et al., 2005).
Likewise, some residues of glutamate dehydrogenase of Peptostreptococcus asaccharolyticus and Clostridium symbiosum has been mutated, due to which catalytic efficiency toward NADPH and NADP+ was increased by several folds, respectively (Griffin and Engel, 2011). Formate dehydrogenase from Candida boidinii (CboFDH) is known to catalyse the oxidation of formate anion to carbon dioxide with concomitant reduction of NAD+ to NADH. It is highly specific to NADH and virtually fails to catalyze the reaction with NADPH, but substitution at Asp195Gln/Tyr196His position causes shift of cofactor specificity from NADH to NADPH (Andreadeli et al., 2008). In isocitrate dehydrogenase from E. coli, site-directed mutagenesis was used to introduce five substitutions (Lys344Asp, Tyr345Ile, Val351Ala, Try391Lys, Arg395Ser) in its adenosine binding pocket that shifts the coenzyme specificity from NADP to NAD (Chen R. et al., 1995). Studies also suggest that a mutation in phenylacetaldehyde dehydrogenase mutation at F180T in lactaldehyde dehydrogenase increases its specificity to NADP+ that also increased enzyme's catalytic activity (Rodríguez-Zavala, 2008).
Same approach was applied to mutate various other enzymes like nitrate reductase, carbonyl reductase etc. which have been highlighted in Table 1(Shiraishi et al., 1998; Morikawa et al., 2005). Microbial production of 1, 3-propanediol has been considered as a competitor to the traditional petrochemical routes. Site directed mutagenesis of 1, 3-propanediol oxidoreductase at Asp48 residue relaxes the cofactor specificity of 1, 3-propanediol toward NADH and NADPH, thereby increasing enzyme efficiency (Ma et al., 2010). NADPH-dependent hydroxybenzoate hydroxylase from Pseudomonas fluorescens was also subjected to site directed mutagenesis that increases its cofactor specificity for NADH. This was the first report on the coenzyme reversion of a flavoprotein aromatic hydroxylase (Eppink et al., 1999).
Change in the catalytic activity of the enzyme
Genetic as well as chemical technologies offer tremendous scope of modifying the basic characteristics of enzymes, e.g., activity, stability, thermal sensitivity etc., to enhance their performance and to make them more suitable for industrial as well as research applications. Aspartate aminotransferases have central role in amino acid metabolism. Out of 13 random mutations, six mutations (Table 1) contributed maximum to the increased activity. Only six amino acids of the enzyme were directly interacting with substrate and results showed that directed molecular evolution is a powerful technique for enzyme redesigning if an adequate selection system is applied (Yano et al., 1998).
Tyrosine phenol lyase of microbial origin from C. freundii uses pyridoxal 5′ phosphate as cofactor and catalyzes the β-elimination of L-tyrosine to produce phenol and ammonium pyruvate. The substitution of Asn185Ala results in formation of a mutant, which has decreased affinity for substrate due to destability of quinonoid structure, formed between them (Barbolina et al., 2000). Similarly, Tyr71Phe was substituted in Tyrosine phenol- lyase and as a result no detectable activity for β-elimination of L-tyrosine was observed and also decreased activity for other substrate was observed (Chen H. Y. et al., 1995). Similarly, Asp124Thr and His448Phe mutations in tryptophan indole-lyase of Proteus vulgaris, have little or no β-elimination activity with L-Tyr or 3-fluoro-L-Tyra substrate, but retain significant elimination activity with S-(o-nitro phenyl)-L-cysteine, S-alkyl-L-cysteines, and β-chloroalanine (Demidkina et al., 2002). In E. coli site-saturation mutagenesis was done and many mutants were made by substituting the active site loop residues. Out of those mutants only some of the substitution mutations increased β–galactosidase activity that is discussed in the Table 1 (Geddie and Matsumura, 2004).
Thermodynamics of the in vitro design of enzyme
In continuation of above discussion, it is also important to study the thermodynamic parameters like ΔG, ΔCp, ΔH involved in the evolutionary path of these enzymes. Using thermodynamic divergence of evolutionary path of mesophilic and thermophilic Ribonuclease H1 from their common ancestor has been traced (Hart et al., 2014). Analyzing its mutants enormous amount of information about the critical positions in enzyme-substrate selectivity can be collected. This is useful in finding their evolution mechanisms i.e., how these enzymes evolved from their ancestors using ancestral sequence reconstruction etc. (Hart et al., 2014). Directed evolution technology to select an enzyme with desired characteristics is one of the earliest and successful methods such as half-life (t1/2) and activity (kcat) of a cold-adapted Lipase B isolated from Candida antarctica were significantly improved from 8 to 211 min and 84 min−1 to 1900 min−1, respectively.
Conclusions
Combined computational and in vitro evolution approaches can be used for the study of evolution of novel bioactivity in the natural enzymes as well as redesigning of the function of the enzyme. DNA shuffling, site-saturation mutagenesis, and site-directed mutagenesis are used can be used to redesign the active site of an enzyme, which can alter its efficiency. Artificial alteration of enzyme can be very useful in changing the catalytic efficiency of enzymes, cofactor specificity, and substrate specificity. Many new enzymes that do not occur naturally can also be developed, which are more efficient. Using these approaches, enzymes can be engineered according to the requirement. For example, if there is defect in any enzyme, the existing related enzyme or isozymes could be engineered in such a way so that it can work in place of defected enzyme. Moreover, the evolutionary relation and thermodynamic system drifts over evolutionary time can also be studied.
Future prospects
Engineered enzymes revolution has been brought about after the discovery of extremophiles, organisms surviving, and thriving in extreme environmental conditions like high temperature, low temperature, high salt, and pH etc. Preliminary studies with extremophile enzymes have provided the researchers vital information about the mechanisms of their adaption of the extreme environments and different catalytic activity. These studies revealed the biochemical and structural features of these enzymes, which facilitated scientists to engineer the mesophilic enzymes to attain thermo- or cryo-stability. Similarly, engineered thermophilic and psychrophilic enzymes attain substrate specificity and activity of mesophilic enzymes and vice versa. For example, thermostable hydrolases (used in starch, food, pharmaceutical, leather, and textile industries), chitinases (used in food, cosmetics, pharmaceuticals, agrochemicals related industries), and phytases (release phosphorus from the phytate are used in animal feed); and cryostable lipases, proteases, phytases, glucanases, and xylanases are used with animal feed; pectinases and xylanases are used in starch hydrolysis at low temperatures; lipases and proteases used in detergent and cleaning industry; lipases are being used in biofuel and energy producing industry etc. Similarly, research activities can also result in the development and production of numerous engineered enzymes with application in industrial processes, therapeutics and household products including food industry, biodiesel, and bioremediation.
Author contributions
Conceived, Designed, Proofread, and corrected: VT.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
VT would like to thanks SERB, DST, India for Start Up grant (SB/YS/LS-07/2014).
References
- Andreadeli A., Platis D., Tishkov V., Popov V., Labrou N. E. (2008). Structure-guided alteration of coenzyme specificity of formate dehydrogenase by saturation mutagenesis to enable efficient utilization of NADP+. FEBS J. 275, 3859–3869. 10.1111/j.1742-4658.2008.06533.x [DOI] [PubMed] [Google Scholar]
- Barbolina M. V., Phillips R. S., Gollnick P. D., Faleev N. G., Demidkina T. V. (2000). Citrobacter freundii tyrosine phenol-lyase: the role of asparagine 185 in modulating enzyme function through stabilization of a quinonoid intermediate. Protein Eng. 13, 207–215. 10.1093/protein/13.3.207 [DOI] [PubMed] [Google Scholar]
- Bell J. K., Yennawar H. P., Wright S. K., Thompson J. R., Viola R. E., Banaszak L. J. (2001). Structural analyses of a malate dehydrogenase with a variable active site. J. Biol. Chem. 276, 31156–31162. 10.1074/jbc.M100902200 [DOI] [PubMed] [Google Scholar]
- Bernard N., Johnsen K., Holbrook J. J., Delcour J. (1995). D175 discriminates between NADH and NADPH in the coenzyme binding site of Lactobacillus delbrueckii subsp. bulgaricus D-lactate dehydrogenase. Biochem. Biophys. Res. Commun. 208, 895–900. 10.1006/bbrc.1995.1419 [DOI] [PubMed] [Google Scholar]
- Brakmann S., Schwienhorst A. (2004). Evolutionary Methods in Biotechnology. Weinheim: Wiley-VCH. [Google Scholar]
- Cahn J. K. B., Baumschlager A., Brinkmann-Chen S., Arnold F. H. (2016). Mutation in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes. Protein Eng. Des. Sel. 29, 31–38. 10.1093/protein/gzv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrigan J. B., Engel P. C. (2007). Probing the determinants of coenzyme specificity in Peptostreptococcus asaccharolyticus glutamate dehydrogenase by site-directed mutagenesis. FEBS J. 274, 5167–5174. 10.1111/j.1742-4658.2007.06038.x [DOI] [PubMed] [Google Scholar]
- Chen H. Y., Demidkina T. V., Phillips R. S. (1995). Site-directed mutagenesis of tyrosine-71 to phenylalanine in Citrobacter freundii tyrosine phenol-lyase: evidence for dual roles of tyrosine-71 as a general acid catalyst in the reaction mechanism and in cofactor binding. Biochemistry 34, 12276–12283. [DOI] [PubMed] [Google Scholar]
- Chen R., Greer A., Dean A. M. (1995). A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity. Proc. Natl. Acad. Sci. U.S.A. 92, 11666–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J. Y., Xue B., Lee K. H., Tung A., Wu L., Robinson R. C., et al. (2010). Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily. J. Biol. Chem. 285, 40911–40920. 10.1074/jbc.M110.177139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti G., Pollegioni L., Molla G., Rosini E. (2014). Strategic manipulation of an industrial biocatalyst-evolution of a cephalosporin C acylase. FEBS J. 281, 2443–2455. 10.1111/febs.12798 [DOI] [PubMed] [Google Scholar]
- Cronin C. N. (1998). Redesign of choline acetyltransferase specificity by protein engineering. J. Biol. Chem. 273, 24465–24469. 10.1074/jbc.273.38.24465 [DOI] [PubMed] [Google Scholar]
- Dembowski N. J., Kantrowitz E. R. (1994). The use of alanine scanning mutagenesis to determine the role of the N-terminus of the regulatory chain in the heterotropic mechanism of Escherichia coli aspartate transcarbamoylase. Protein Eng. 7, 673–679. 10.1093/protein/7.5.673 [DOI] [PubMed] [Google Scholar]
- Demidkina T. V., Barbolina M. V., Faleev N. G., Sundararaju B., Gollnick P. D., Phillips R. S. (2002). Threonine-124 and phenylalanine-448 in Citrobacter freundii tyrosine phenol-lyase are necessary for activity with L-tyrosine. Biochem. J. 363, 745–752. 10.1042/bj3630745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. (1973). Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 35, 125–129. 10.2307/4444260 [DOI] [Google Scholar]
- Eppink M. H., Overkamp K. M., Schreuder H. A., Van Berkel W. J. (1999). Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase. J. Mol. Biol. 292, 87–96. 10.1006/jmbi.1999.3015 [DOI] [PubMed] [Google Scholar]
- Flores H., Ellington A. D. (2005). A modified consensus approach to mutagenesis inverts the cofactor specificity of Bacillus stearothermophilus lactate dehydrogenase. Protein Eng. Des. Sel. 18, 369–377. 10.1093/protein/gzi043 [DOI] [PubMed] [Google Scholar]
- Geddie M. L., Matsumura I. (2004). Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop. J. Biol. Chem. 279, 26462–26468. 10.1074/jbc.M401447200 [DOI] [PubMed] [Google Scholar]
- Ge Y. D., Song P., Cao Z. Y., Wang P., Zhu G. P. (2014). Alteration of coenzyme specificity of malate dehydrogenase from Streptomyces coelicolor A3(2) by site-directed mutagenesis. Genet. Mol. Res. 13, 5758–5766. 10.4238/2014.July.29.3 [DOI] [PubMed] [Google Scholar]
- Goh K. M., Honga G. P., Chyi N. H., Piawa C. K., Rahman R. N. Z. R. A. (2012). Trends and tips in protein engineering, a review. Jurnal Teknologi (Sci. Eng.) 59, 21–31. [Google Scholar]
- Graber R., Kasper P., Malashkevich V. N., Strop P., Gehring H., Jansonius J. N., et al. (1999). Conversion of aspartate aminotransferase into an L-aspartate beta-decarboxylase by a triple active-site mutation. J. Biol. Chem. 274, 31203–31208. 10.1074/jbc.274.44.31203 [DOI] [PubMed] [Google Scholar]
- Graycar T., Knapp M., Ganshaw G., Dauberman J., Bott R. (1999). Engineered Bacillus lentus subtilisins having altered flexibility. J. Mol. Biol. 292, 97–109. 10.1006/jmbi.1999.3033 [DOI] [PubMed] [Google Scholar]
- Griffin J., Engel P. C. (2011). An examination by site-directed mutagenesis of putative key residues in the determination of coenzyme specificity in clostridial NAD-dependent glutamate dehydrogenase. Enzyme Res. 2011:595793. 10.4061/2011/595793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber K., Klintschar G., Hayn M., Schlacher A., Steiner W., Kratky C. (1998). Thermophilic xylanase from thermomyces lanuginosus: high resolution X-ray structure and modelling study. Biochemistry 37, 13475–13485. 10.1021/bi980864l [DOI] [PubMed] [Google Scholar]
- Hall B. G., Barlow M. (2004). Evolution of the serine beta-lactamases: past, present and future. Drug Resist. Updat. 7, 111–123. 10.1016/j.drup.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Hart K. M., Harms M. J., Schmidt B. H., Elya C., Thornton J. W., Marqusee S. (2014). Thermodynamic system drift in protein evolution. PLoS Biol. 11:e1001994. 10.1371/journal.pbio.1001994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Nakayama K. (2016). Alteration of substrate selection of antibiotic acylase from beta-lactam to echinocandin. Protein Eng. Des. Sel. 29, 49–56. 10.1093/protein/gzv059 [DOI] [PubMed] [Google Scholar]
- Kaushik M., Sinha P., Jaiswal P., Mahendru S., Roy K., Kukreti S. (2016). Protein engineering and de novo designing of a biocatalyst. J. Mol. Recognit. 29, 499–503. 10.1002/jmr.2546 [DOI] [PubMed] [Google Scholar]
- Kawata T., Ogino H. (2010). Amino acid residues involved in organic solvent-stability of the LST-03 lipase. Biochem. Biophys. Res. Commun. 400, 384–388. 10.1016/j.bbrc.2010.08.080 [DOI] [PubMed] [Google Scholar]
- Leemhuis H., Kelly R. M., Dijkhuizen L. (2010). Engineering of cyclodextrin glucanotransferases and the impact for biotechnological applications. Appl. Microbiol. Biotechnol. 85, 823–835. 10.1007/s00253-009-2221-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cirino P. C. (2014). Recent advances in engineering proteins for biocatalysis. Biotechnol. Bioeng. 111, 1273–1287. 10.1002/bit.25240 [DOI] [PubMed] [Google Scholar]
- Ma C., Zhang L., Dai J., Xiu Z. (2010). Relaxing the coenzyme specificity of 1,3-propanediol oxidoreductase from Klebsiella pneumoniae by rational design. J. Biotechnol. 146, 173–178. 10.1016/j.jbiotec.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Madern D., Cai X., Abrahamsen M. S., Zhu G. (2004). Evolution of Cryptosporidium parvum lactate dehydrogenase from malate dehydrogenase by a very recent event of gene duplication. Mol. Biol. Evol. 21, 489–497. 10.1093/molbev/msh042 [DOI] [PubMed] [Google Scholar]
- Morikawa S., Nakai T., Yasohara Y., Nanba H., Kizaki N., Hasegawa J. (2005). Highly active mutants of carbonyl reductase S1 with inverted coenzyme specificity and production of optically active alcohols. Biosci. Biotechnol. Biochem. 69, 544–552. 10.1271/bbb.69.544 [DOI] [PubMed] [Google Scholar]
- Morimoto Y., Honda K., Ye X., Okano K., Ohtake H. (2014). Directed evolution of thermotolerant malic enzyme for improved malate production. J. Biosci. Bioeng. 117, 147–152. 10.1016/j.jbiosc.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Mouratou B., Kasper P., Gehring H., Christen P. (1999). Conversion of tyrosine phenol-lyase to dicarboxylic amino acid beta-lyase, an enzyme not found in nature. J. Biol. Chem. 274, 1320–1325. 10.1074/jbc.274.3.1320 [DOI] [PubMed] [Google Scholar]
- Nishiyama M., Birktoft J. J., Beppu T. (1993). Alteration of coenzyme specificity of malate dehydrogenase from Thermus flavus by site-directed mutagenesis. J. Biol. Chem. 268, 4656–4660. [PubMed] [Google Scholar]
- Nyon M. P., Rice D. W., Berrisford J. M., Hounslow A. M., Moir A. J., Huang H., et al. (2009). Catalysis by Glomerella cingulata cutinase requires conformational cycling between the active and inactive states of its catalytic triad. J. Mol. Biol. 385, 226–235. 10.1016/j.jmb.2008.10.050 [DOI] [PubMed] [Google Scholar]
- Oue S., Okamoto A., Yano T., Kagamiyama H. (1999). Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. J. Biol. Chem. 274, 2344–2349. 10.1074/jbc.274.4.2344 [DOI] [PubMed] [Google Scholar]
- Petschacher B., Leitgeb S., Kavanagh K. L., Wilson D. K., Nidetzky B. (2005). The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem. J. 385, 75–83. 10.1042/BJ20040363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski B. T., Buckle A. M. (2016). Consensus protein design. Protein Eng. Des. Sel. 29, 245–251. 10.1093/protein/gzw015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik G. O., Vajda S., Sano T., Cantor C. R. (1998). A streptavidin mutant with altered ligand-binding specificity. Proc. Natl. Acad. Sci. U.S.A. 95, 13525–13530. 10.1073/pnas.95.23.13525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Zavala J. S. (2008). Enhancement of coenzyme binding by a single point mutation at the coenzyme binding domain of E. coli lactaldehyde dehydrogenase. Protein Sci. 17, 563–570. 10.1110/ps.073277108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld L., Heyne M., Shifman J. M., Papo N. (2016). protein engineering by combined computational and in vitro evolution approaches. Trends Biochem. Sci. 41, 421–433. 10.1016/j.tibs.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Shiraishi N., Croy C., Kaur J., Campbell W. H. (1998). Engineering of pyridine nucleotide specificity of nitrate reductase: mutagenesis of recombinant cytochrome b reductase fragment of Neurospora crassa NADPH:Nitrate reductase. Arch. Biochem. Biophys. 358, 104–115. 10.1006/abbi.1998.0827 [DOI] [PubMed] [Google Scholar]
- Tokuda C., Ishikura Y., Shigematsu M., Mutoh H., Tsuzuki S., Nakahira Y., et al. (2003). Conversion of Lactobacillus pentosus D-lactate dehydrogenase to a D-hydroxyisocaproate dehydrogenase through a single amino acid replacement. J. Bacteriol. 185, 5023–5026. 10.1128/JB.185.16.5023-5026.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T., Fushinobu S., Kuzuyama T., Nishiyama M. (2006a). Structural basis for the alteration of coenzyme specificity in a malate dehydrogenase mutant. Biochem. Biophys. Res. Commun. 347, 502–508. 10.1016/j.bbrc.2006.06.131 [DOI] [PubMed] [Google Scholar]
- Tomita T., Kuzuyama T., Nishiyama M. (2006b). Alteration of coenzyme specificity of lactate dehydrogenase from Thermus thermophilus by introducing the loop region of NADP(H)-dependent malate dehydrogenase. Biosci. Biotechnol. Biochem. 70, 2230–2235. 10.1271/bbb.60170 [DOI] [PubMed] [Google Scholar]
- Toyama M., Yamashita M., Yoneda M., Zaborowski A., Nagato M., Ono H., et al. (2002). Alteration of substrate specificity of cholesterol oxidase from Streptomyces sp. by site-directed mutagenesis. Protein Eng. 15, 477–484. 10.1093/protein/15.6.477 [DOI] [PubMed] [Google Scholar]
- Volkov A. A., Arnold F. H. (2000). Methods for in vitro DNA recombination and random chimeragenesis. Methods Enzymol. 328, 447–456. 10.1016/S0076-6879(00)28411-5 [DOI] [PubMed] [Google Scholar]
- Wright S. K., Kish M. M., Viola R. E. (2000). From malate dehydrogenase to phenyllactate dehydrogenase. Incorporation of unnatural amino acids to generate an improved enzyme-catalyzed activity. J. Biol. Chem. 275, 31689–31694. 10.1074/jbc.M003652200 [DOI] [PubMed] [Google Scholar]
- Wright S. K., Viola R. E. (2001). Alteration of the specificity of malate dehydrogenase by chemical modulation of an active site arginine. J. Biol. Chem. 276, 31151–31155. 10.1074/jbc.M100892200 [DOI] [PubMed] [Google Scholar]
- Wu G., Fiser A., Ter Kuile B., Sali A., Müller M. (1999). Convergent evolution of Trichomonas vaginalis lactate dehydrogenase from malate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 96, 6285–6290. 10.1073/pnas.96.11.6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T., Oue S., Kagamiyama H. (1998). Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc. Natl. Acad. Sci. U.S.A. 95, 5511–5515. 10.1073/pnas.95.10.5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. X., Hui R., Lin M., Qing H. G., Jun T. X., He C. Q. (2006). Enhancement of the thermostability of β- 1,3-1,4-glucanase by directed evolution. J. Zhejiang Univ. Sci. A 7, 1948–1955. 10.1631/jzus.2006.A1948 [DOI] [Google Scholar]
- Zeng Q. K., Du H. L., Wang J. F., Wei D. Q., Wang X. N., Li Y. X., et al. (2009). Reversal of coenzyme specificity and improvement of catalytic efficiency of Pichia stipitis xylose reductase by rational site-directed mutagenesis. Biotechnol. Lett. 31, 1025–1029. 10.1007/s10529-009-9980-x [DOI] [PubMed] [Google Scholar]