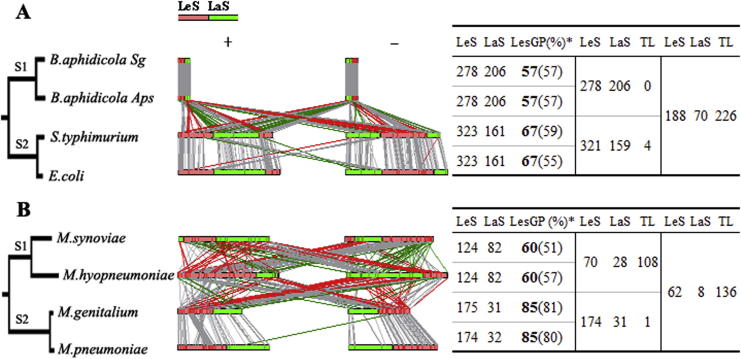
Figure 3.
A case study on strand-preference of conserved genes of the non-polC and polC bacteria We classified the strand distribution of 484 conserved genes in Buchnera (non-polC group; A) and 206 conserved genes in Mycoplasma (polC group; B). We built phylogenetic trees based on the NJ method (bootstrap value = 1000) using 16s rRNA sequences (Mega 4.0; the left panel). The trees were drawn to the scale, with branch lengths in the same units as those of the evolutionary distances used to infer phylogeny. We computed the evolutionary distance using the number of nucleotide variations per sequence as the unit. We selected two bacteria, Ehrlichia canis and Mesoplasma florum, as outgroups to root the trees in (A) and (B), respectively. All positions containing gaps and missing data were eliminated from the dataset. Genes on the positive (+) and negative (-) strands were separated into the LeS (solid red bars) and LaS (solid green bars) groups (the middle panel) and the direction of gene transfer were color-coded accordingly: LaS to LeS (red), LeS to LaS (green), no inter-strand transfer event (gray). The number of genes and their distributions were summarized in the table (the right panel). LesGP values (%) of conserved (bold) and total genes (in parentheses) were also calculated. Genes transferring between strands are classified as translocatable (TL).