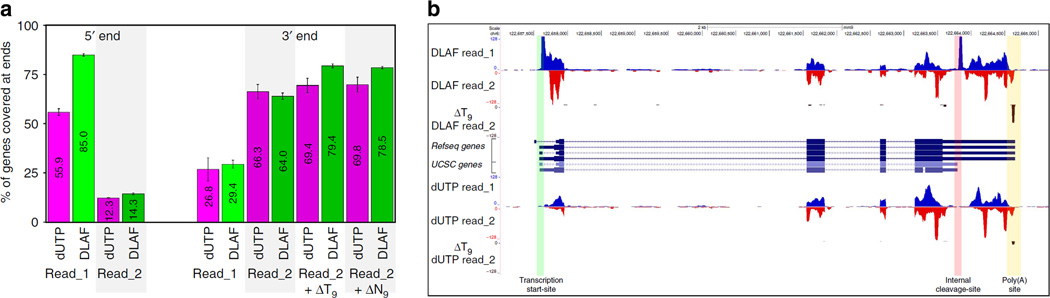
Figure 6. DLAF results in end-to-end coverage of transcriptome.
(a) Percentage of genes covered at 5′ and 3′ ends. RNA-SeQC data are shown for the 2,500 middle-expressed genes in WT mES cells. Data are shown for 12.5 million randomly selected non-rRNA and non-mtRNA reads. Average of two biological replicates is shown and error-bars indicate the range of data. (b) Image from the UCSC genome browser of the Nanog locus. DLAF read_1 shows a distinct coverage at the annotated TSS (green border) and an internal CS (pink border). Remapping of DLAF read_2 after ΔT9 analysis identified the polyadenylation site (yellow border). Signals are normalized to the total number of non-rRNA and non-mtRNA reads from each library.