Figure 2.
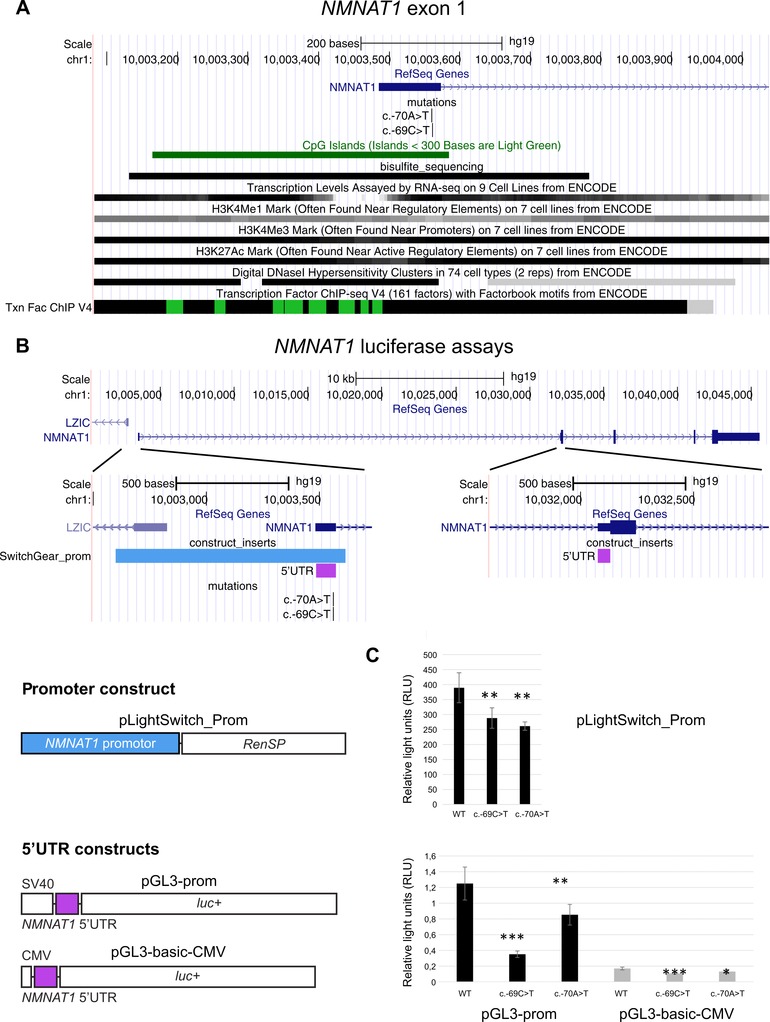
Location and functional analysis of the NMNAT1 5′UTR mutations, c.‐70A>T and c.‐69C>T. A: Overview of a selection of the ENCODE Integrated Regulation tracks aligning with exon 1. The bisulfite_sequencing custom track shows the region covered by the bisulfite sequencing performed in F1. B: Overview of the NMNAT1 gene locus and luciferase reporter constructs. The NMNAT1 5′UTR consists of exon 1 and part of exon 2, which are separated from each other by a large intron (28 kb). The promoter construct contains the NMNAT1 promoter upstream of a Renilla luciferase gene. The 5′UTR constructs contain an SV40/CMV promoter, followed by the NMNAT1 5′UTR and a firefly luciferase gene. C: Results luciferase assays. Upper panel: transcriptional activities of WT and mutated NMNAT1 promoter. Luciferase assays were performed in RPE‐1 cells. Relative lights units (RLU) correspond to the ratio of the activity of the Renilla reporter (RenSP) over that of the firefly luciferase reporter (internal control of transfection efficiency, pGL3‐CMV‐empty). Lower panel: transcriptional activities of WT and mutated NMNAT1 5′UTR. RLU correspond to the ratio of the activity of the firefly luciferase reporter over that of the Renilla reporter (internal control of transfection efficiency, pRL‐RSV). Statistical significance in Student's t‐tests: n.s., P.0.05; *: P,0.05; **, P,0.01, ***, P,0.001.
