Figure 3.
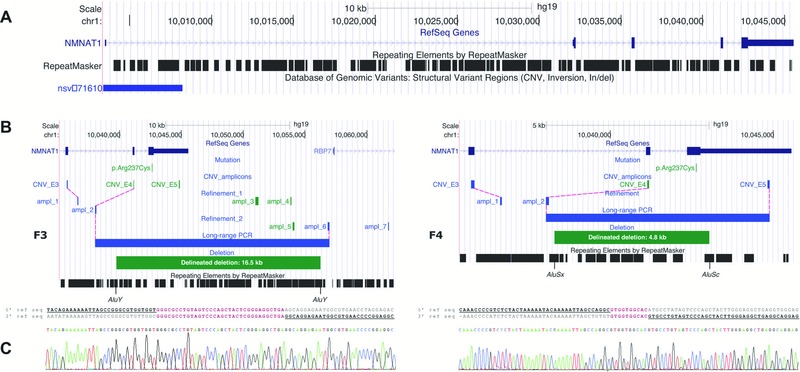
Molecular characterization of two distinct heterozygous NMNAT1 deletions. A: Overview of the NMNAT1 genomic locus, which is scattered with Alu repeats. The Database of Genomic Variants contains one duplication. B: Refinement and delineation of both heterozygous deletions at the nucleotide level. Deleted amplicons are indicated in green; amplicons with a normal copy number are indicated in blue. In F3, two subsequent refinement rounds with seven additional qPCR amplicons in total (ampl_1–ampl_7) were needed to refine the deletion at both ends to a region of 13.0–18.7 kb. Subsequent long‐range PCR and Sanger sequencing revealed a deletion of 16.5 kb. In F4, refinement at the 5′ end (ampl_1 and ampl_2) delineated the deletion to a region of 1.5–6.7 kb. At the 3′ end, the deletion breakpoint appeared to be located between the p.Arg237Cys mutation and the forward primer of the amplicon CNV_E5 (3′ UTR), as this amplicon was not deleted according to qPCR results (Supp. Fig. S6B). Subsequent long‐range PCR and Sanger sequencing revealed a deletion of 4.8 kb. For both deletions, the breakpoints regions are located in Alu repeats. C: Alignment of the junction product with the 5′ and 3′ reference sequence revealed microhomology regions of 34 and 10 bp for F3 and F4, respectively.
