Figure 4.
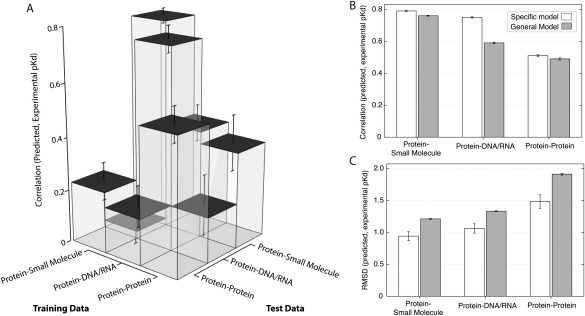
Binding affinity prediction is specific to each interaction type. For each type of molecular interaction (protein‐small molecule, protein‐DNA/RNA, and protein−protein), we fit a statistical model using cross‐validation (see Methods, Fig. 2) and evaluated each model's accuracy on set‐aside testing data of either the same interaction type or a different interaction type. A: We plot the mean and standard error in Pearson's correlation (r 2) between predicted and experimental pKd, showing how the statistical models trained using each type of training data predicted pKds on testing data of either the same or different type. B: We plot the mean and standard error in r 2 between predicted and experimental pKd, comparing a general statistical model trained on all data sets (gray) to specific models (white) trained on each data set, respectively. C: We show the mean and standard error in root mean square deviation (RMSD) between predicted and experimental pKd, comparing a general statistical model (gray) trained on all data sets to specific models (white) trained on each data set.
