Figure 7.
Nascent Chain Ribosome Complexes Accumulate on Early Assembly Factors upon Block of Complex IV Assembly
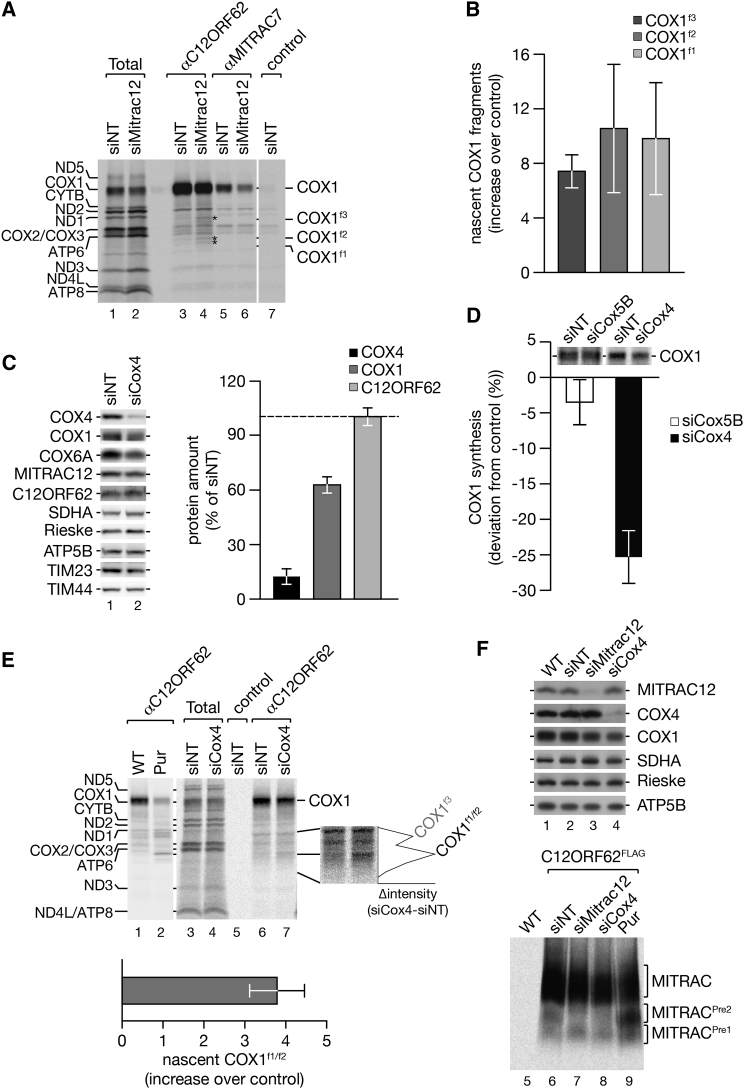
(A and B) MITRAC12 was depleted in HEK293T cells by siRNA treatment for 72 hr prior to [35S]methionine labeling and immunoisolation using antibodies as indicated (A). COX1 fragments are labeled by asterisks. Total, 5%; eluates, 100%. (B) Quantification of COX1 fragments upon MITRAC12 depletion (mean ± SEM; n = 3).
(C) Cell lysates (50 μg for MITRAC12, C12ORF62, SDHA, and TIM44; 25 μg for all other proteins) were analyzed by SDS-PAGE and western blotting. Quantification of COX4, COX1, and C12ORF62 protein levels (mean ± SEM, n = 4) after 72 hr knock-down of COX4.
(D) Quantification of newly synthesized COX1 upon COX5B and COX4 depletion. Level of COX1 synthesis (standardized to ATP6) were calculated and normalized to non-targeting control (mean ± SEM; n = 3).
(E) COX4 was ablated in HEK293T cells prior to radiolabeling of mitochondrial translation products. Samples were subjected to immunoprecipitation using indicated antibodies. Radiolabeled protein bands were quantified along the gel lanes and the differential profile (Δintensity) plotted along the gel section of interest. Bottom graph, increase of COX1f1/f2 fragments relative to the control (mean ± SEM; n = 4).
(F) COX4 and MITRAC12 were depleted in HEK293T cells expressing C12ORF62FLAG by siRNA treatment for 72 hr prior to [35S]methionine labeling and FLAG-immunoisolation. Upper panel: western blot analysis of cell lysates. Lower panel: BN-PAGE analysis of natively eluted immunoisolation samples. As a control, one sample was treated with puromycin prior to isolation omitting siRNA treatment.
See also Figure S5.