Figure 2.
Chromatin and Intracellular Transport Dominate Strong Metabolic Signatures
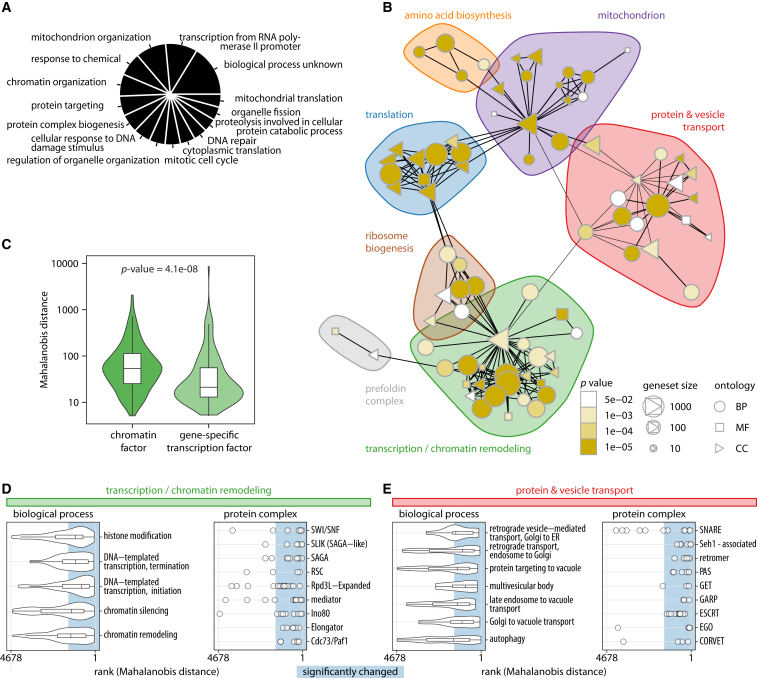
(A) Top 15 Gene Ontology (GO) slim categories of gene deletions affecting the biosynthetic metabolome.
(B) Functional “perturbation network” constructed on the basis of a gene set enrichment analysis (GSEA). Each node represents an enriched GO term (adjusted p value < 0.05), which is connected when the gene overlap is >50%. Edges are drawn for shortest paths of one or two; distinct groups were identified by clustering of the edge betweenness. BP, biological process; MF, molecular function; CC, cellular component. A fully labeled version is given in Figure S2.
(C) Chromatin- and histone-modifying proteins, as part of the general gene expression machinery, have a significantly stronger impact than gene-specific transcription factors (Wilcoxon rank-sum test).
(D) Protein complexes associated with general transcriptional control at the chromatin level trigger strong metabolic signatures (4,678: most similar to wild-type; 1: most different to wild-type).
(E) Protein complexes associated with vesicle-mediated transport processes that are responsible for strong metabolic signatures (4,678: most similar to wild-type; 1: most different to wild-type).